Simulating the Normal-Normal in RJAGS
Bayesian Modeling with RJAGS

Alicia Johnson
Associate Professor, Macalester College
Sleep study
$Y_i$ = change in reaction time (ms) after 3 days of sleep deprivation
$Y_i \sim N(m, s^2)$
Insights from the priors
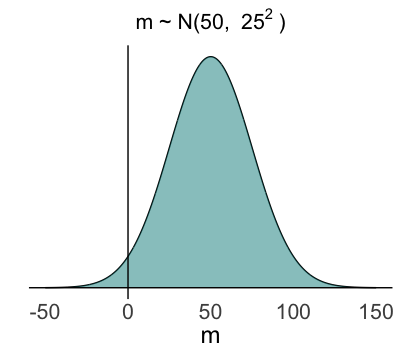
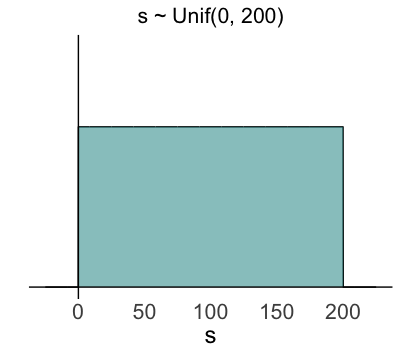
Insights from the data (& likelihood)
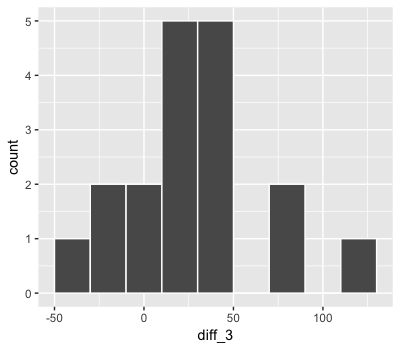
mean(sleep_study$diff_3)
sd(sleep_study$diff_3)
26.34021
37.20764
Assuming these data are generated from $Y_i \sim N(m, s^2)$, they are most likely to have occurred if...
- $m \approx 26$ ms
- $s \approx 37$ ms
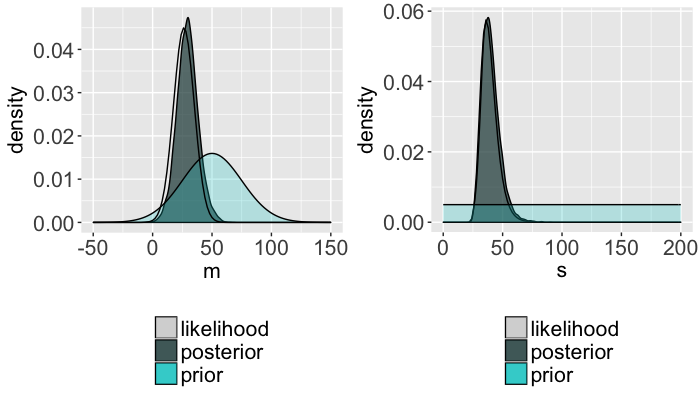
Posterior insights

DEFINE the Normal-Normal
sleep_model <- "model{
# Likelihood model for Y[i]
# Prior models for m and s
}"
DEFINE the Normal-Normal
sleep_model <- "model{
# Likelihood model for Y[i]
# Prior models for m and s
}"
- $Y_i \sim N(m, s^2)$ for $i$ in ${1,2,\ldots,18}$
DEFINE the Normal-Normal
sleep_model <- "model{
# Likelihood model for Y[i]
for(i in 1:length(Y)) {
}
# Prior models for m and s
}"
- $Y_i \sim N(m, s^2)$ for $i$ in ${1,2,\ldots,18}$
DEFINE the Normal-Normal
sleep_model <- "model{
# Likelihood model for Y[i]
for(i in 1:length(Y)) {
Y[i] ~ dnorm(m, s^(-2))
}
# Prior models for m and s
}"
- $Y_i \sim N(m, s^2)$ for $i$ in ${1,2,\ldots,18}$
- NOTE: precision = variance$^{-1}$ = $s^{-2}$
DEFINE the Normal-Normal
sleep_model <- "model{
# Likelihood model for Y[i]
for(i in 1:length(Y)) {
Y[i] ~ dnorm(m, s^(-2))
}
# Prior models for m and s
m ~ dnorm(50, 25^(-2))
}"
- $Y_i \sim N(m, s^2)$ for $i$ in ${1,2,\ldots,18}$
- NOTE: precision = variance$^{-1}$ = $s^{-2}$
- $m \sim N(50, 25^2)$
DEFINE the Normal-Normal
sleep_model <- "model{
# Likelihood model for Y[i]
for(i in 1:length(Y)) {
Y[i] ~ dnorm(m, s^(-2))
}
# Prior models for m and s
m ~ dnorm(50, 25^(-2))
s ~ dunif(0, 200)
}"
- $Y_i \sim N(m, s^2)$ for $i$ in ${1,2,\ldots,18}$
- NOTE: precision = variance$^{-1}$ = $s^{-2}$
- $m \sim N(50, 25^2)$
- $s \sim \text{Unif}(0, 200)$
COMPILE the Normal-Normal
# COMPILE the model
sleep_jags <- jags.model(textConnection(sleep_model),
data = list(Y = sleep_study$diff_3),
inits = list(.RNG.name = "base::Wichmann-Hill",
.RNG.seed = 1989))
sleep_study$diff_3
[1] 71.8798 -18.0269 33.7877 -36.4096 32.5074 74.9082
[7] 15.9673 -10.8008 29.1938 33.7556 18.8188 -0.7697
[13] 30.0626 125.1784 5.7331 15.2090 11.9091 41.2199
SIMULATE the Normal-Normal
# COMPILE the model
sleep_jags <- jags.model(textConnection(sleep_model),
data = list(Y = sleep_study$diff_3),
inits = list(.RNG.name = "base::Wichmann-Hill",
.RNG.seed = 1989))
# SIMULATE the posterior
sleep_sim <- coda.samples(model = sleep_jags,
variable.names = c("m", "s"),
n.iter = 10000)
SIMULATE the Normal-Normal
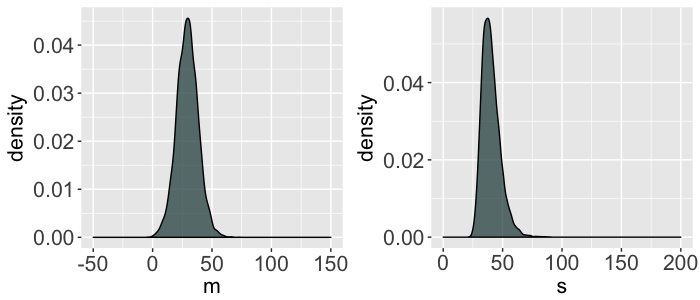
SIMULATE the Normal-Normal
Let's practice!
Bayesian Modeling with RJAGS

