The posterior model
Bayesian Modeling with RJAGS

Alicia Johnson
Associate Professor, Macalester College
Bayesian election model
prior: $p \sim \text{Beta}(45, 55)$
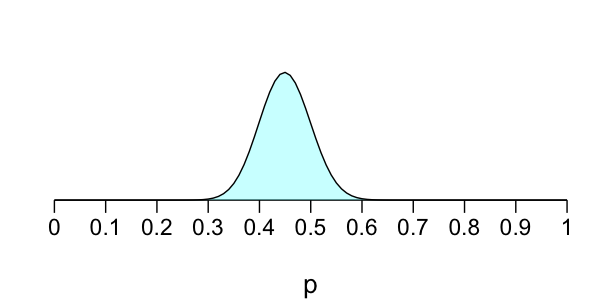
Bayesian election model
Prior: $p \sim \text{Beta}(45, 55)$
Likelihood: $X \sim \text{Beta}(10, p)$
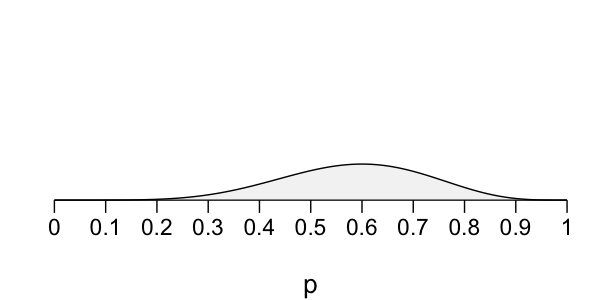
Bayesian election model
Prior: $p \sim \text{Beta}(45, 55)$
Likelihood: $X \sim \text{Bin}(10, p)$
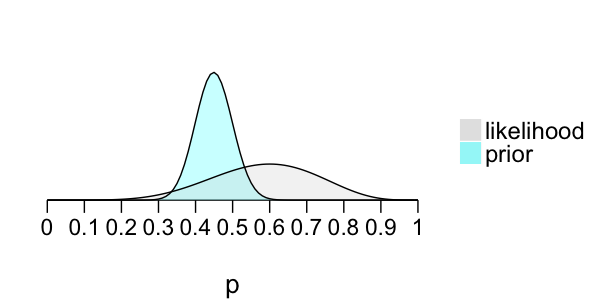
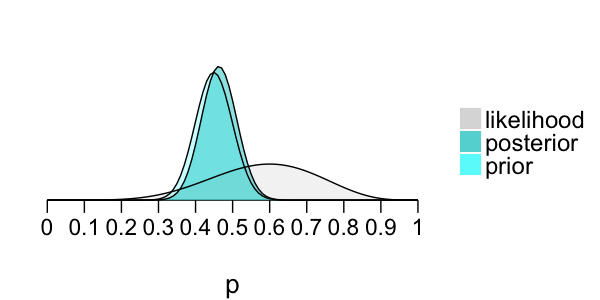
Posterior model of p
Prior: $p \sim \text{Beta}(45, 55)$
Likelihood: $X \sim \text{Bin}(10, p)$
Bayes' Rule:
posterior $\propto$ prior $\times$ likelihood
Getting started with RJAGS
RJAGS combines the power of R with the JAGS (Just Another Gibbs Sampler) engine. To get started:
- Download the
JAGSprogram outsideR - Within
R, install therjagspackage
Bayesian models in RJAGS: DEFINE
# DEFINE the model
vote_model <- "model{
# Likelihood model for X
X ~ dbin(p, n)
# Prior model for p
p ~ dbeta(a, b)
}"
- $X \sim \text{Bin}(n, p)$
- $p \sim \text{Beta}(a, b)$
- Warning:
therjagsfunctiondbin()is different than basedbinom()
Bayesian models in RJAGS: COMPILE
# DEFINE the model
vote_model <- "model{
# Likelihood model for X
X ~ dbin(p, n)
# Prior model for p
p ~ dbeta(a, b)
}"
# COMPILE the model
vote_jags_A <- jags.model(textConnection(vote_model),
data = list(a = 45, b = 55, X = 6, n = 10),
inits = list(.RNG.name = "base::Wichmann-Hill", .RNG.seed = 100))
Bayesian models in RJAGS: SIMULATE
# DEFINE the model
vote_model <- "model{
# Likelihood model for X
X ~ dbin(p, n)
# Prior model for p
p ~ dbeta(a, b)
}"
# COMPILE the model
vote_jags <- jags.model(textConnection(vote_model),
data = list(a = 45, b = 55, X = 6, n = 10),
inits = list(.RNG.name = "base::Wichmann-Hill", .RNG.seed = 100))
# SIMULATE the posterior
vote_sim <- coda.samples(model = vote_jags,
variable.names = c("p"),
n.iter = 10000)
Bayesian models in RJAGS: SIMULATE
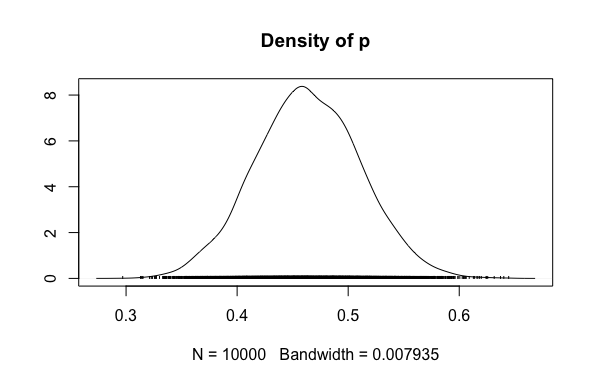
# PLOT the simulated posterior
plot(vote_sim, trace = FALSE)
Let's practice!
Bayesian Modeling with RJAGS

