Differential expression data
Differential Expression Analysis with limma in R

John Blischak
Instructor
The experimental data
Study of breast cancer
- Bioconductor package "breastCancerVDX"
- Published in Wang et al., 2005 and Minn et al., 2007
- 344 patients: 209 ER+, 135 ER-
Study of chronic lymphocytic leukemia (CLL)
- Bioconductor package "CLL"
- Drs. Sabina Chiaretti and Jerome Ritz
- 22 patients: 8 stable, 14 progressive
Data in R
Expression matrix (
x)Feature data (
f) - feature attributesPhenotype data (
p) - sample attributes
Expression matrix
rows = features, columns = samples
class(x)
"matrix"
dim(x)
22283 344
x[1:5, 1:5]
VDX_3 VDX_5 VDX_6
1007_s_at 11.965135 11.798593 11.777625
1053_at 7.895424 7.885696 7.949535
117_at 8.259272 7.052025 8.225930
Feature data
rows = features, columns = any number of attributes
class(f)
"data frame"
dim(f)
22283 3
f[1:3, ]
symbol entrez chrom
1007_s_at DDR1 780 6p21.3
1053_at RFC2 5982 7q11.23
117_at HSPA6 3310 1q23
Phenotype data
rows = samples, columns = any number of attributes
class(p)
"data frame"
dim(p)
344 3
# er = +/- for Estrogen Receptor
p[1:3, ]
id age er
VDX_3 3 36 negative
VDX_5 5 47 positive
VDX_6 6 44 negative
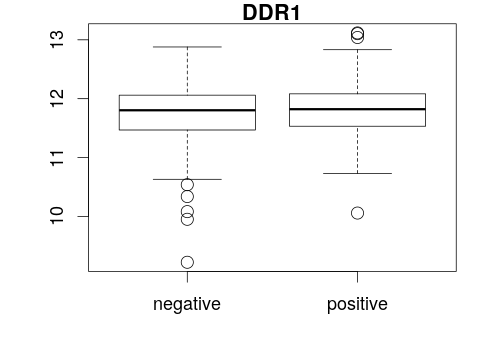
Visualize gene expression with a boxplot
boxplot(<y-axis> ~ <x-axis>, main = "<title>")boxplot(<gene expression> ~ <phenotype>, main = "<feature>")boxplot(x[1, ] ~ p[, "er"], main = f[1, "symbol"])
Let's practice!
Differential Expression Analysis with limma in R

