The ExpressionSet class
Differential Expression Analysis with limma in R

John Blischak
Instructor
Data management is precarious
x_sub <- x[1000, 1:10]
f_sub <- f[1000, ]
p_sub <- p[1:10, ]
A single misplaced comma could become a debugging nightmare:
x_sub <- x[1000, 1:10]
f_sub <- f[1000, ]
p_sub <- p[, 1:10]
# Oh no! *
Object-oriented programming with Bioconductor classes
class - defines a structure to hold complex data
object - a specific instance of a class
methods - functions that work on a specific class
- getters/accessors - Get data stored in an object
- setters/ - Modify data stored in an object
install.packages("BiocManager")
BiocManager::install("Biobase")
Create an ExpressionSet object
# Load package library(Biobase) # Create ExpressionSet object eset <- ExpressionSet(assayData = x, phenoData = AnnotatedDataFrame(p), featureData = AnnotatedDataFrame(f))# View the number of features (rows) and samples (columns) dim(eset)
Features Samples
22283 344
?ExpressionSet
Access data from an ExpressionSet object
Expression matrix
x <- exprs(eset)
Feature data
f <- fData(eset)
Phenotype data
p <- pData(eset)
Subset an ExpressionSet object
- Subset with 3 separate objects:
x_sub <- x[1000, 1:10]
f_sub <- f[1000, ]
p_sub <- p[1:10, ]
- Subset with an ExpressionSet object:
eset_sub <- eset[1000, 1:10]
nrow(exprs(eset_sub)) == nrow(fData(eset_sub))
TRUE
ncol(exprs(eset_sub)) == nrow(pData(eset_sub))
TRUE
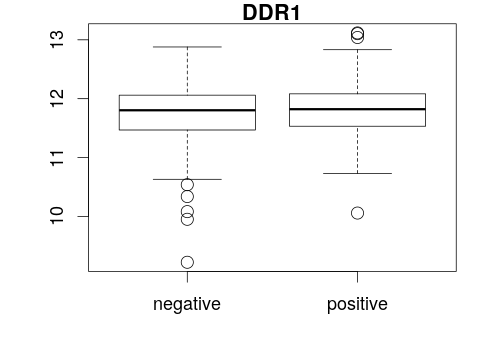
Boxplot with an ExpressionSet
boxplot(<y-axis> ~ <x-axis>, main = "<title>") boxplot(<gene expression> ~ <phenotype>, main = "<feature>")boxplot(exprs(eset)[1, ] ~ pData(eset)[, "er"], main = fData(eset)[1, "symbol"])
Let's practice!
Differential Expression Analysis with limma in R

