Visualizing the results
Differential Expression Analysis with limma in R

John Blischak
Instructor
Inspecting the results
results <- decideTests(fit2)
summary(results)
topTable(fit2, number = 3)
status
-1 6276
0 11003
1 5004
symbol entrez chrom logFC AveExpr t
205225_at ESR1 2099 6q25.1 3.762901 11.37774 22.68392
209603_at GATA3 2625 10p15 3.052348 9.94199 18.98154
209604_s_at GATA3 2625 10p15 2.431309 13.18533 17.59968
P.Value adj.P.Val B
205225_at 2.001001e-70 4.458832e-66 149.1987
209603_at 1.486522e-55 1.656209e-51 115.4641
209604_s_at 5.839050e-50 4.337052e-46 102.7571
Obtain results for all genes
stats <- topTable(fit2, number = nrow(fit2), sort.by = "none")
dim(stats)
22283 9
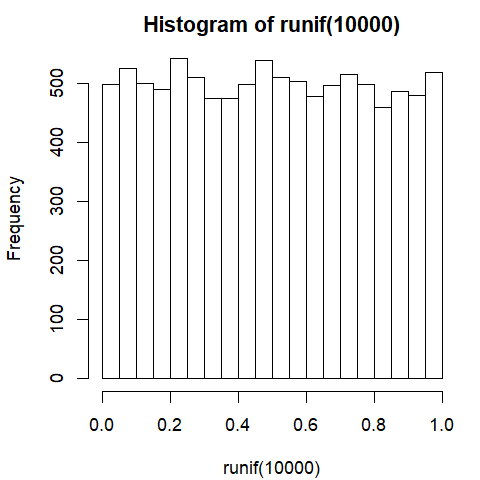
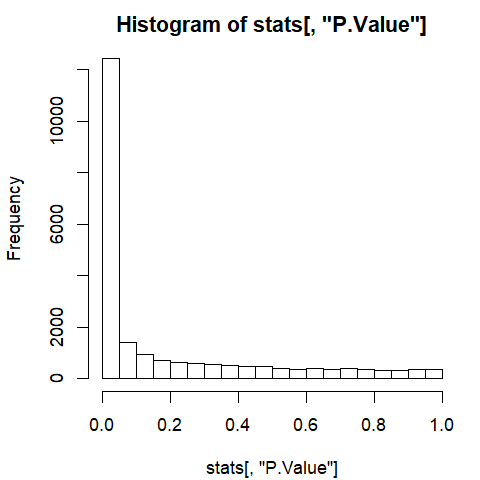
Histogram of p-values
hist(runif(10000))
hist(stats[, "P.Value"])
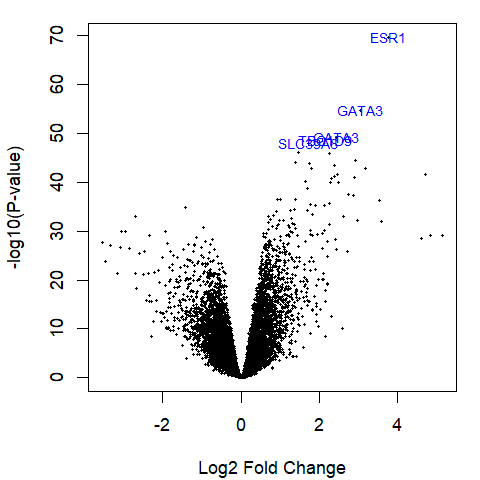
Volcano plot
volcanoplot(fit2,
highlight = 5,
names = fit2$genes[, "symbol"])
Let's practice!
Differential Expression Analysis with limma in R

