Normalizing and filtering
Differential Expression Analysis with limma in R

John Blischak
Instructor
Pre-processing steps
Log transform
Quantile normalize
Filter
Visualization
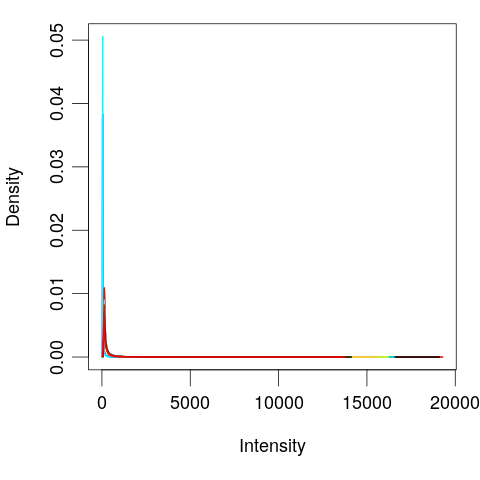
library(limma)
# Plot distribution of each sample
plotDensities(eset, legend = FALSE)
Log transform
100 - 1
.1 - .001
99
0.099
log(100) - log(1)
4.60517
log(.1) - log(.001)
4.60517
# Log tranform
exprs(eset) <- log(exprs(eset))
plotDensities(eset, legend = FALSE)
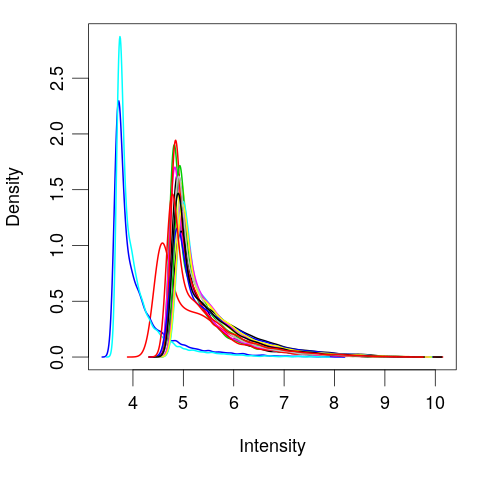
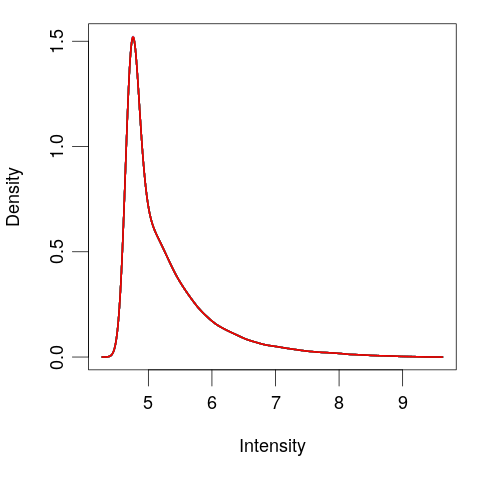
Quantile normalize
# Quantile normalize exprs(eset) <- normalizeBetweenArrays(exprs(eset))plotDensities(eset, legend = FALSE)
Filter genes
# View the normalized data
plotDensities(eset, legend = FALSE)
abline(v = 5)
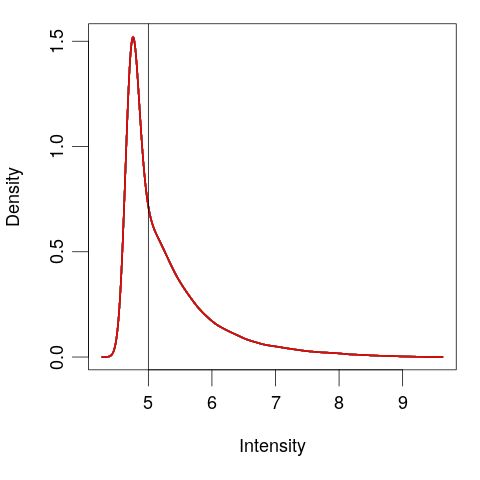
# Create logical vector
keep <- rowMeans(exprs(eset)) > 5
# Filter the genes
eset <- eset[keep, ]
plotDensities(eset, legend = FALSE)
Let's practice!
Differential Expression Analysis with limma in R

