Model the data
Differential Expression Analysis with limma in R

John Blischak
Instructor
Ready for analysis
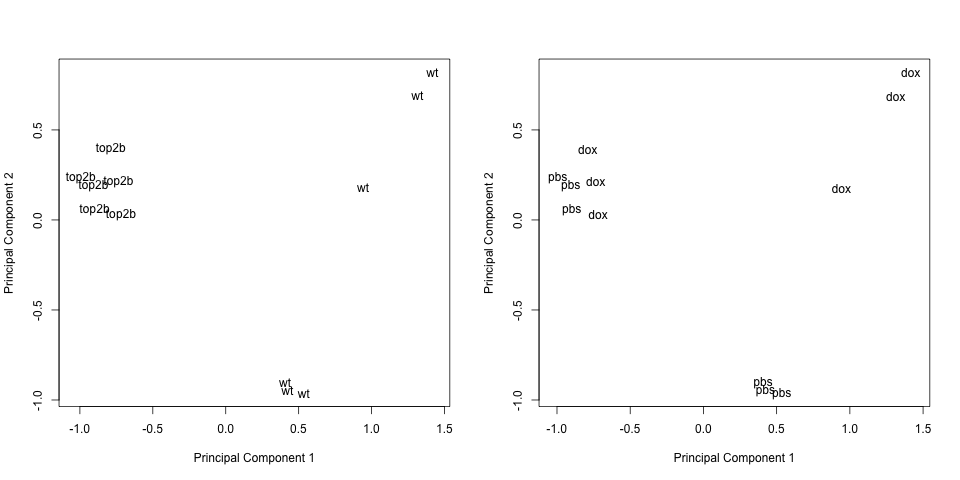
# Plot principal components labeled by genotype
plotMDS(eset, labels = pData(eset)[, "genotype"], gene.selection = "common")
# Plot principal components labeled by treatment
plotMDS(eset, labels = pData(eset)[, "treatment"], gene.selection = "common")
Steps for differential expression analysis
Build the design matrix with
model.matrixContruct the contrasts matrix with
makeContrastsTest the contrasts with
lmFit,contrasts.fit, andeBayes
Group-means model for doxorubicin study
$$ Y = \beta_1 X_1 + \beta_2 X_2 + \beta_3 X_3 + \beta_4 X_4 + \epsilon $$
- $\beta_1$ - Mean expression level in
top2bmice treated withdox - $\beta_2$ - Mean expression level in
top2btreated withpbs - $\beta_3$ - Mean expression level in
wtmice treated withdox - $\beta_4$ - Mean expression level in
wtmice treated withpbs
Contrasts for doxorubicin study
| $\beta_1$ | $\beta_2$ | $\beta_3$ | $\beta_4$ | |
|---|---|---|---|---|
genotype |
top2b | top2b | wt | wt |
treatment |
dox | pbs | dox | pbs |
Response of wild type mice to dox treatment: $\beta_3 - \beta_4 = 0$
Response of Top2b null mice to dox treatment: $\beta_1 - \beta_2 = 0$
Differences between Top2b null and wild type mice in response to dox treatment: $(\beta_1 - \beta_2) - (\beta_3 - \beta_4) = 0$
Testing the doxorubicin study
Fit the model coefficients with
lmFitFit the contrasts with
contrasts.fitCalculate the t-statistics with
eBayes
# Summarize results
results <- decideTests(fit2)
summary(results)
# Create a Venn diagram
vennDiagram(results)
Let's practice!
Differential Expression Analysis with limma in R

