Pre-process the data
Differential Expression Analysis with limma in R

John Blischak
Instructor
Mechanism of doxorubicin-induced cardiotoxicity
- 2x2 design to study mechanism of doxorubicin:
- 2 genotypes: wild type (
wt), Top2b null (top2b) - 2 treatments: PBS (
pbs), doxorubicin (dox) - Zhang et al. 2012
- 2 genotypes: wild type (
dim(eset)
Features Samples
29532 12
table(pData(eset)[, c("genotype", "treatment")])
treatment
genotype dox pbs
top2b 3 3
wt 3 3
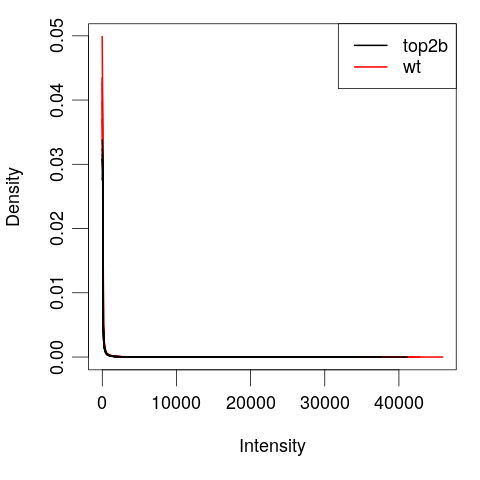
Inspect the features
plotDensities(eset,
group = pData(eset)[, "genotype"],
legend = "topright")
Pre-processing steps
Log transform
Quantile normalize
Filter
Sanity check: Boxplot of Top2b
boxplot(<y-axis> ~ <x-axis>, main = "<title>")
boxplot(<gene expression> ~ <phenotype>, main = "<feature>")
boxplot(<Top2b expression> ~ <genotype>, main = "<Top2b info>")
Check sources of variation
Principal components analysis
limma function
plotMDSHow many clusters do you anticipate?
Let's practice!
Differential Expression Analysis with limma in R

