Visualization of results
RNA-Seq with Bioconductor in R

Mary Piper
Bioinformatics Consultant and Trainer
Visualizing results - Expression heatmap
# Subset normalized counts to significant genes sig_norm_counts_wt <- normalized_counts_wt[wt_res_sig$ensgene, ]# Choose a color palette from RColorBrewer library(RColorBrewer) heat_colors <- brewer.pal(6, "YlOrRd")display.brewer.all()
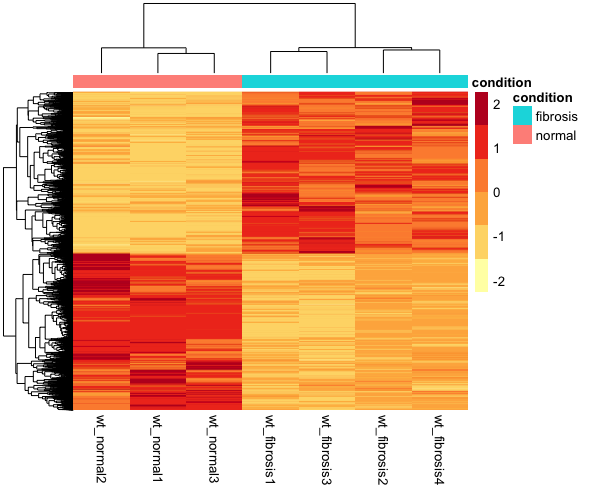
Visualizing results - Expression heatmap
# Run pheatmap
pheatmap(sig_norm_counts_wt,
color = heat_colors,
cluster_rows = T,
show_rownames = F,
annotation = select(wt_metadata, condition),
scale = "row")
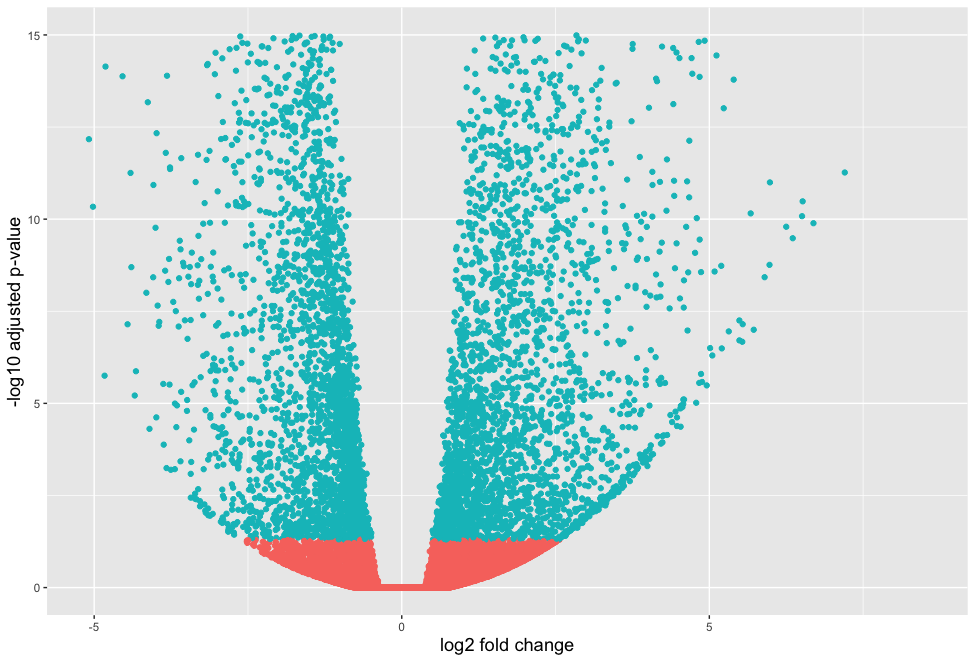
Visualizing results - Volcano plot
# Obtain logical vector regarding whether padj values are less than 0.05 wt_res_all <- wt_res_all %>% rownames_to_column(var = "ensgene") %>% mutate(threshold = padj < 0.05)# Volcano plot ggplot(wt_res_all) + geom_point(aes(x = log2FoldChange, y = -log10(padj), color = threshold)) + xlab("log2 fold change") + ylab("-log10 adjusted p-value") + theme(legend.position = "none", plot.title = element_text(size = rel(1.5), hjust = 0.5), axis.title = element_text(size = rel(1.25)))
ggplot(wt_res_all) +
geom_point(aes(x = log2FoldChange, y = -log10(padj), color = threshold)) +
xlab("log2 fold change") +
ylab("-log10 adjusted p-value") +
ylim=c(0, 15) +
theme(legend.position = "none",
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = rel(1.25)))
Visualizing results - Expression plot
Significant results:
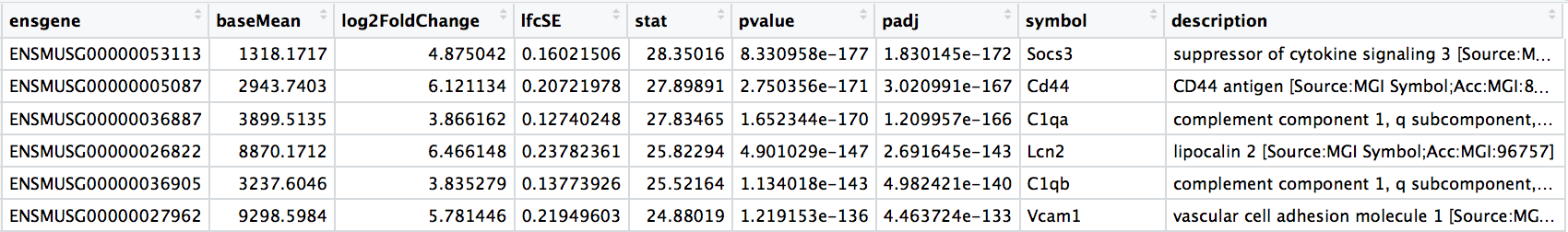
Normalized counts for significant genes:
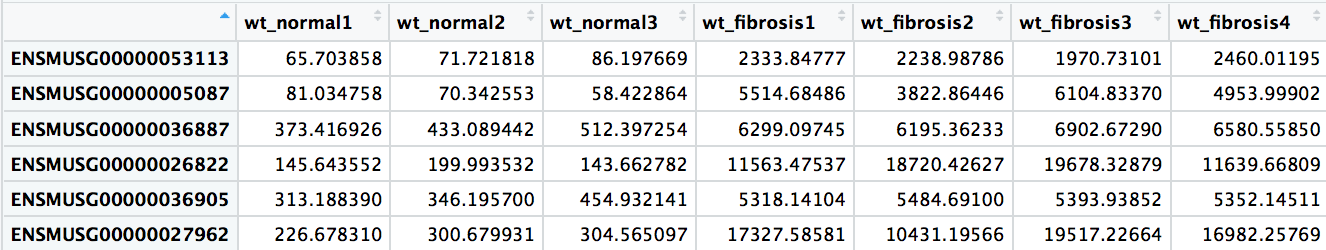
Visualizing results - Expression plot
top_20 <- data.frame(sig_norm_counts_wt)[1:20, ] %>% rownames_to_column(var = "ensgene")top_20 <- gather(top_20, key = "samplename", value = "normalized_counts", 2:8)
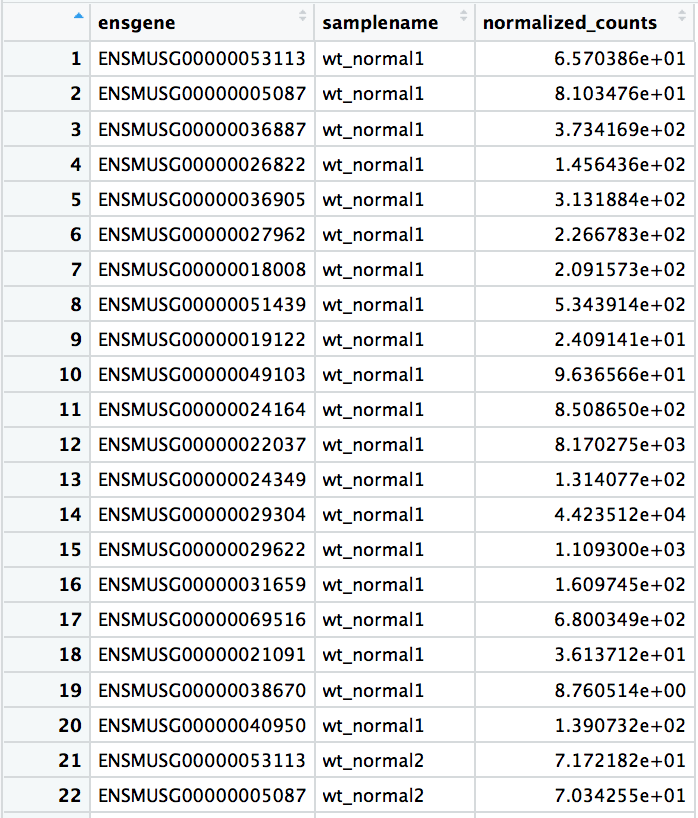
Visualizing results - Expression plot
top_20 <- inner_join(top_20,
rownames_to_column(wt_metadata, var = "samplename"),
by = "samplename")
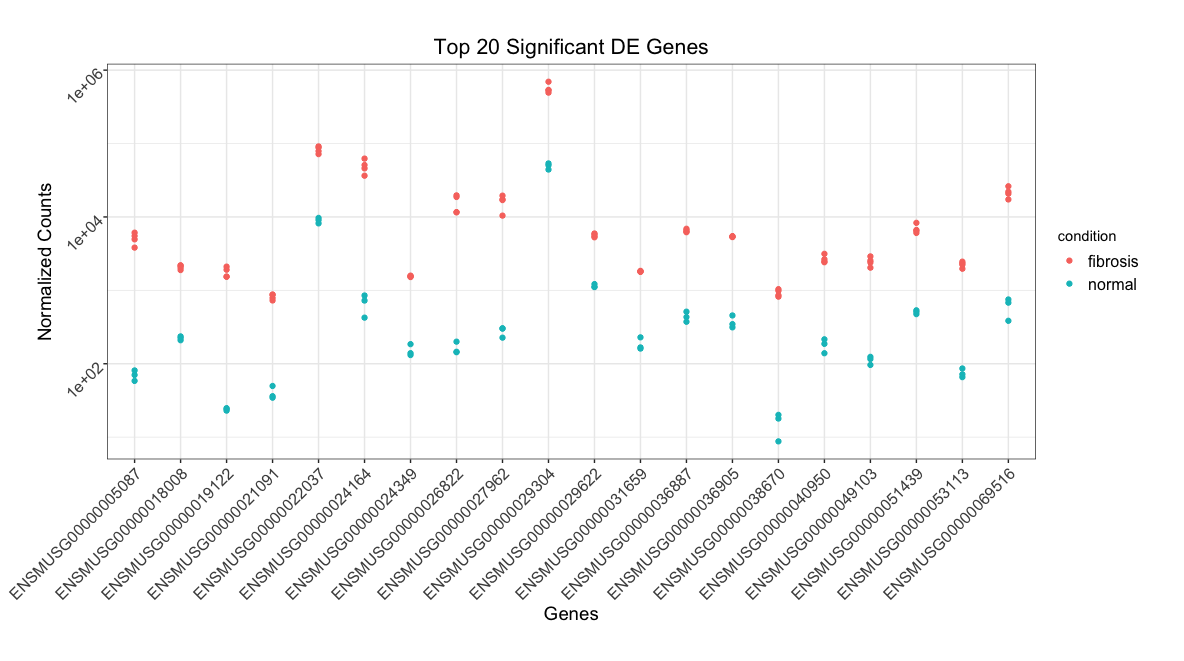
ggplot(top_20) +
geom_point(aes(x = ensgene, y = normalized_counts, color = condition)) +
scale_y_log10() +
xlab("Genes") +
ylab("Normalized Counts") +
ggtitle("Top 20 Significant DE Genes") +
theme_bw() +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
theme(plot.title = element_text(hjust = 0.5))
Let's practice!
RNA-Seq with Bioconductor in R

