DESeq2 model - contrasts
RNA-Seq with Bioconductor in R

Mary Piper
Bioinformatics Consultant and Trainer
DESEq2 workflow
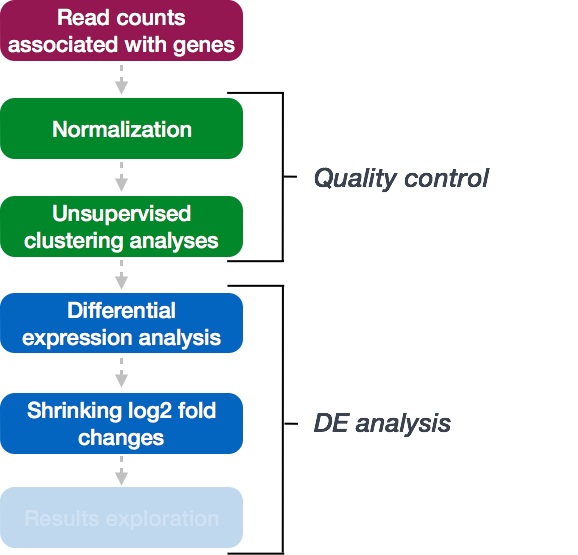
DESeq2 workflow
# Run analysis
dds_wt <- DESeq(dds_wt)
using pre-existing size factors
estimating dispersions
gene-wise dispersion estimates
mean-dispersion relationship
final dispersion estimates
fitting model and testing
DESeq2 Negative Binomial Model
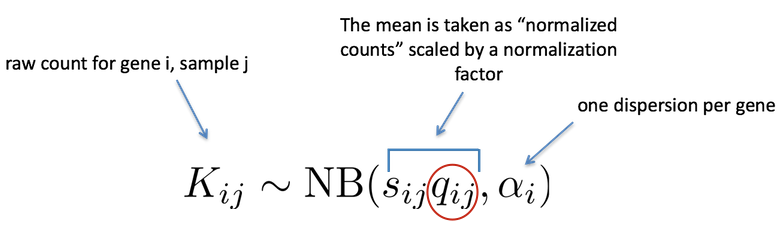
DESeq2 Negative Binomial Model
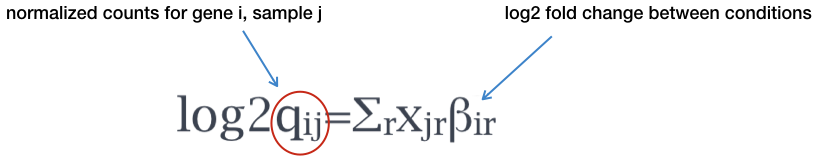
DESeq2 contrasts
results(wt_dds, alpha = 0.05)
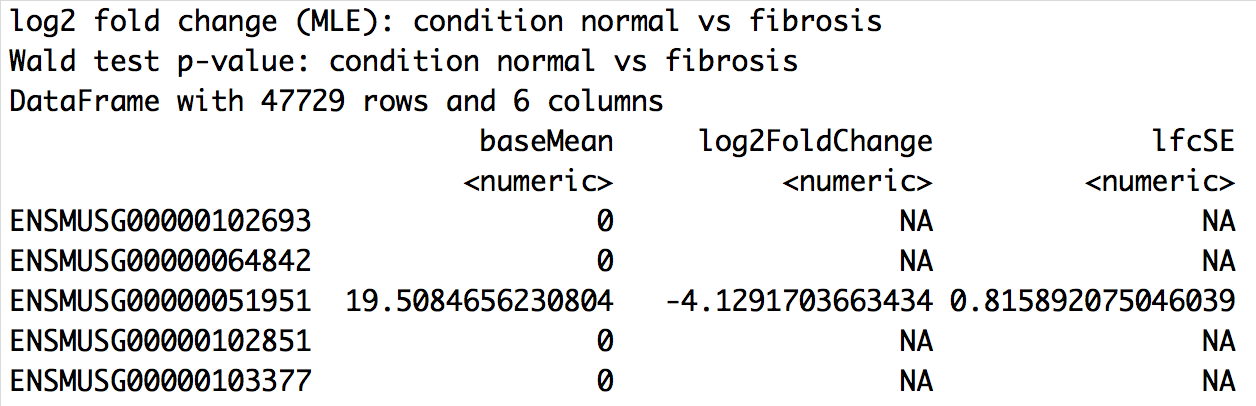
DESeq2 contrasts
The syntax is:
results(dds,
contrast = c("condition_factor", "level_to_compare",
"base_level"),
alpha = 0.05)
wt_res <- results(dds_wt,
contrast = c("condition", "fibrosis",
"normal"),
alpha = 0.05)
DESeq2 contrasts
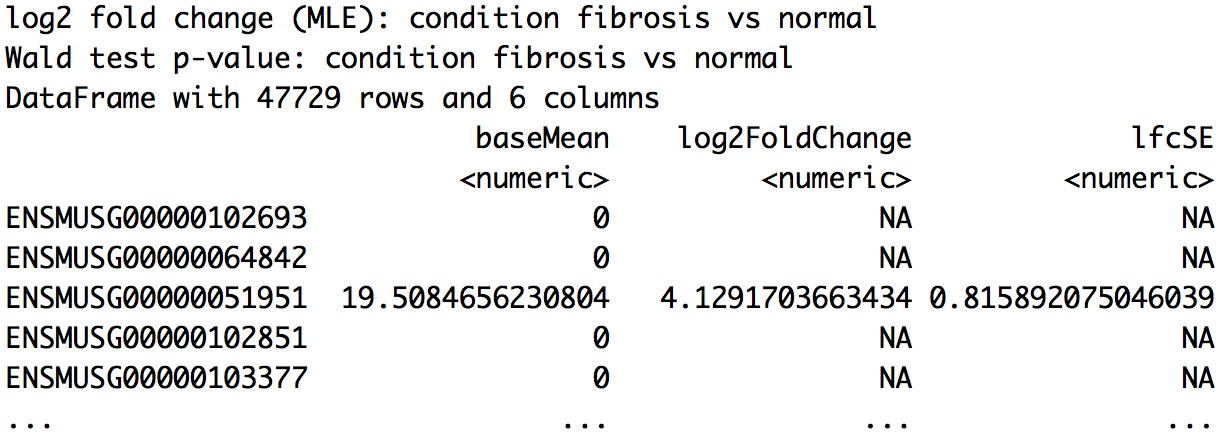
wt_res
DESeq2 LFC shrinkage
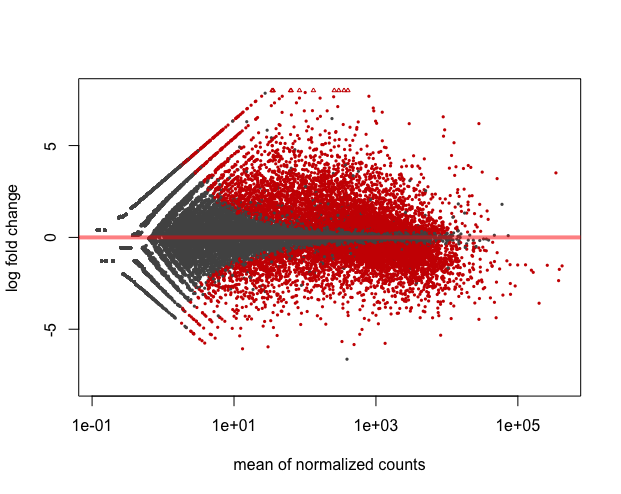
plotMA(wt_res, ylim=c(-8,8))
LFC shrinkage
wt_res <- lfcShrink(dds_wt,
contrast=c("condition", "fibrosis", "normal"),
res=wt_res)
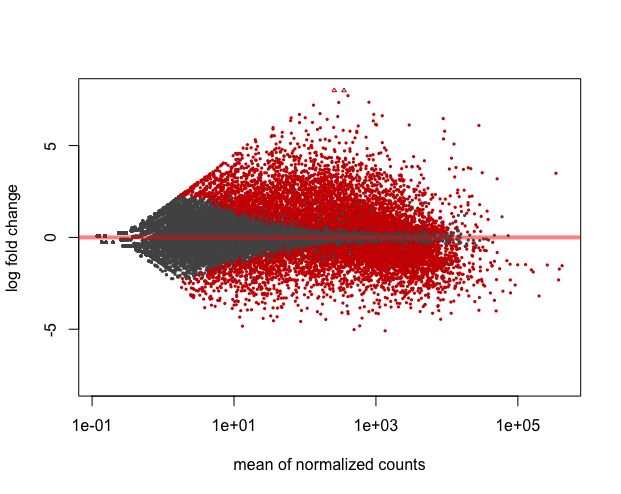
plotMA(wt_res, ylim=c(-8,8))
LFC shrinkage
Let's practice!
RNA-Seq with Bioconductor in R

