DESeq2 model
RNA-Seq with Bioconductor in R

Mary Piper
Bioinformatics Consultant and Trainer
DESeq2 model
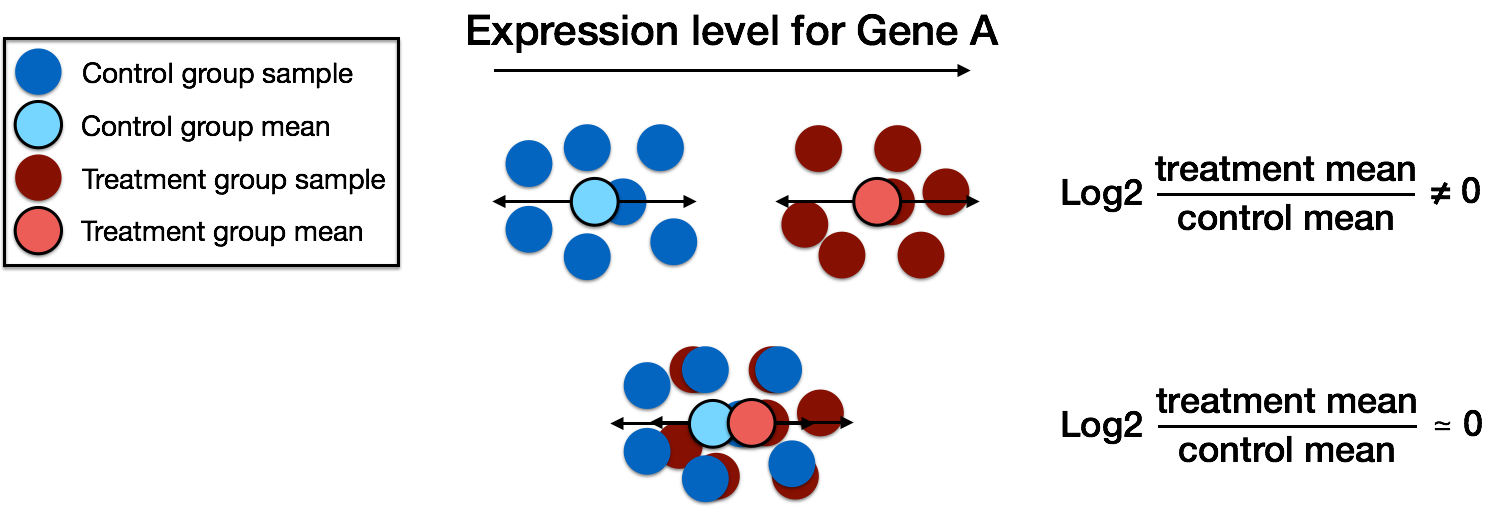
DESeq2 model - mean-variance relationship
# Syntax for apply()
apply(data, rows/columns, function_to_apply)
# Calculating mean for each gene (each row)
mean_counts <- apply(wt_rawcounts[, 1:3], 1, mean)
# Calculating variance for each gene (each row)
variance_counts <- apply(wt_rawcounts[, 1:3], 1, var)
DESeq2 model - dispersion
Plotting relationship between mean and variance:
# Creating data frame with mean and variance for every gene
df <- data.frame(mean_counts, variance_counts)
ggplot(df) +
geom_point(aes(x=mean_counts, y=variance_counts)) +
scale_y_log10() +
scale_x_log10() +
xlab("Mean counts per gene") +
ylab("Variance per gene")
DESeq2 model - dispersion
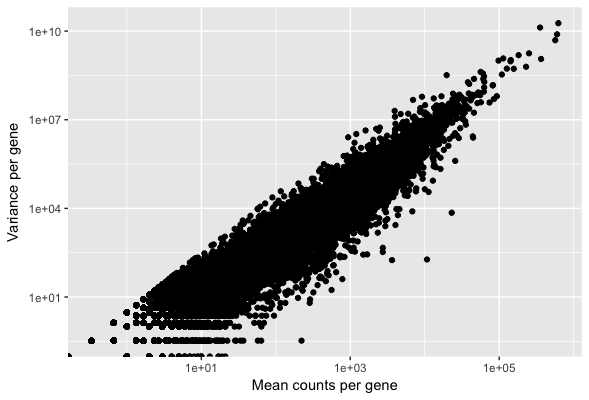
DESeq2 model - dispersion
$Var$: variance
$\mu$: mean
$\alpha$: dispersion
Dispersion formula: $Var = \mu + \alpha * \mu^{2}$
Relationship between mean, variance and dispersion:
$$\uparrow variance \Rightarrow \uparrow dispersion$$
$$\uparrow mean \Rightarrow \downarrow dispersion$$
DESeq2 model - dispersion
# Plot dispersion estimates
plotDispEsts(dds_wt)
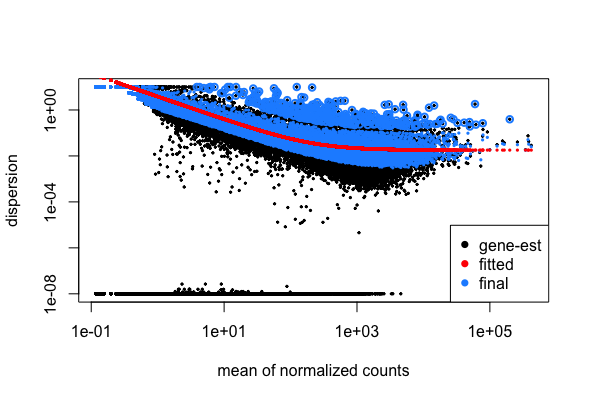
DESeq2 model - dispersion
Let's practice!
RNA-Seq with Bioconductor in R

