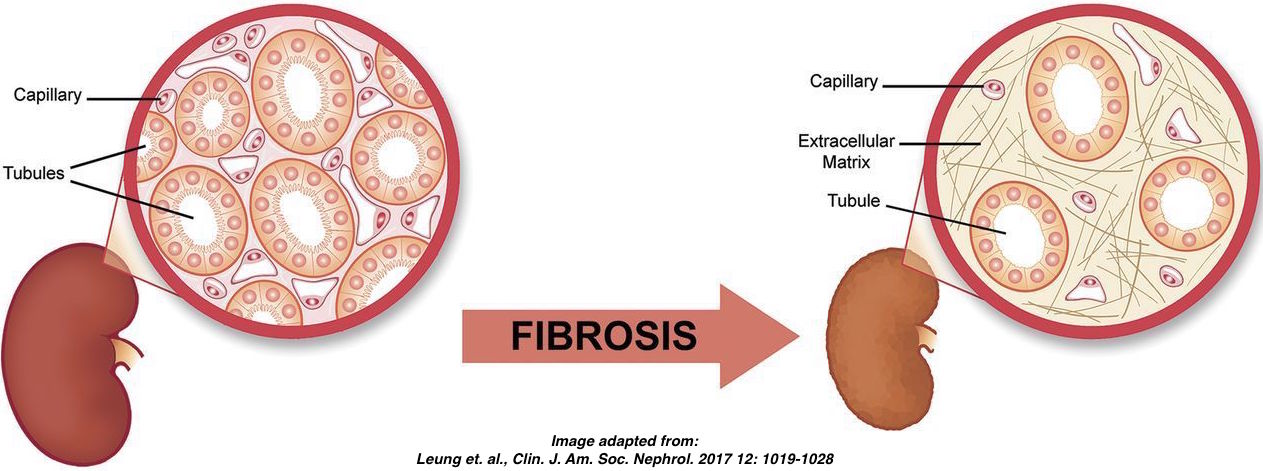
Overview of the DE analysis
RNA-Seq with Bioconductor in R

Mary Piper
Bioinformatics Consultant and Trainer
Review the dataset/question
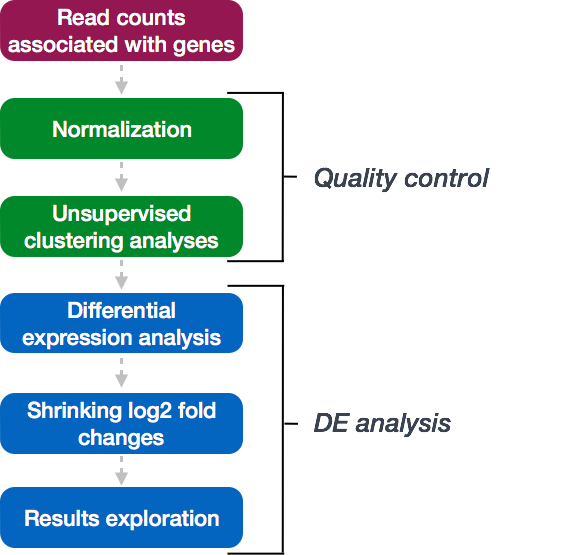
Overview of the DE analysis
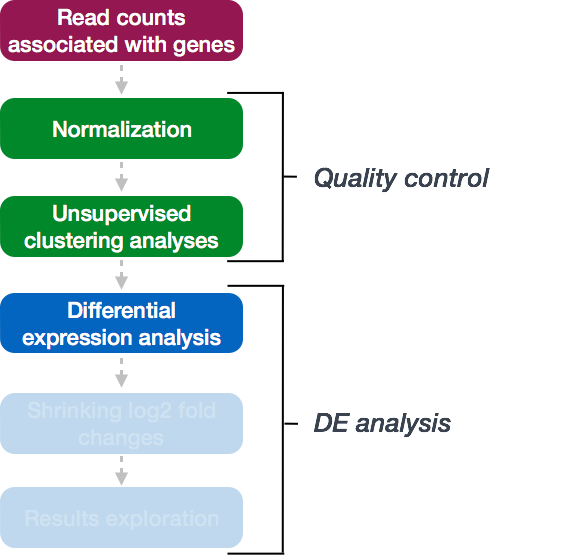
DESeq2 workflow: Model
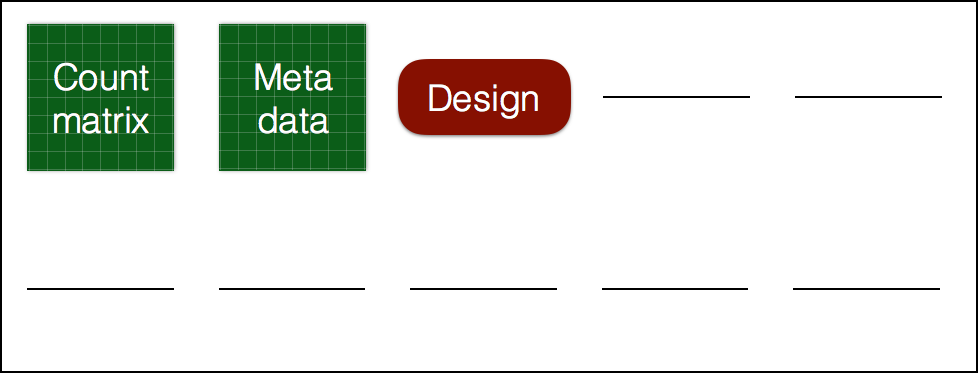
# Create DESeq object
dds_wt <- DESeqDataSetFromMatrix(countData = wt_rawcounts,
colData = reordered_wt_metadata,
design = ~ condition)
DESeq2 workflow: Design formula
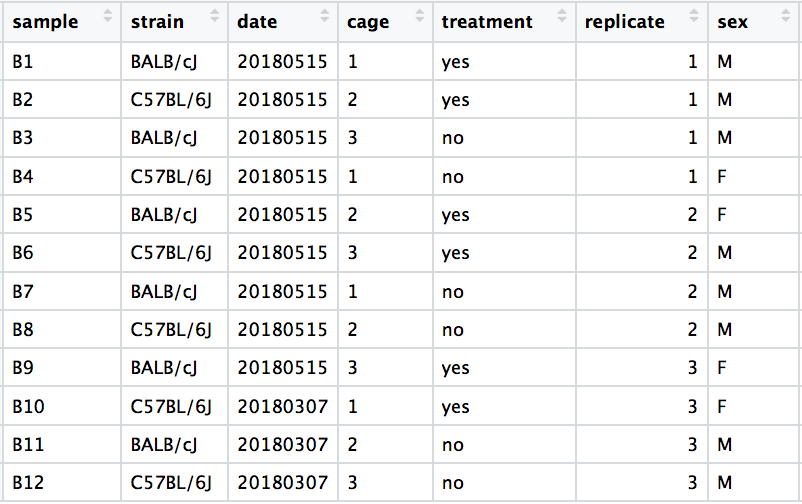
# Design formula
~ strain + sex + treatment
DESeq2 workflow: Design formula

# Design formula
~ strain + sex + treatment + sex:treatment
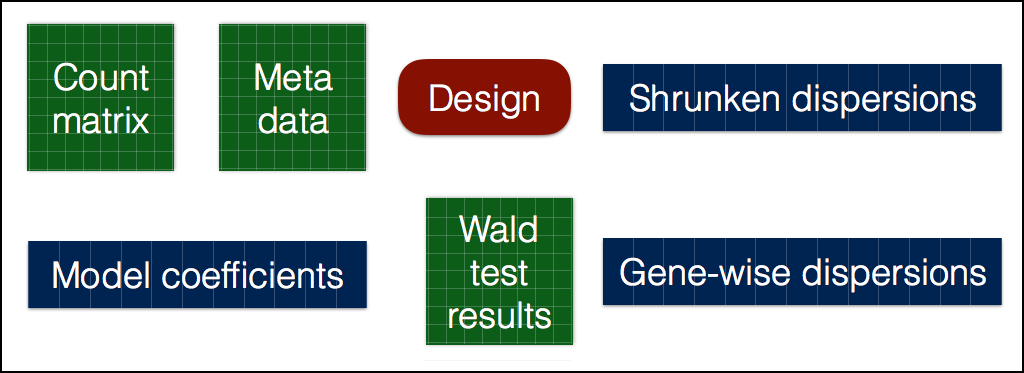
DESeq2 workflow: Running
# Run analysis
dds_wt <- DESeq(dds_wt)
using pre-existing size factors
estimating dispersions
gene-wise dispersion estimates
mean-dispersion relationship
final dispersion estimates
fitting model and testing
Let's practice!
RNA-Seq with Bioconductor in R

