Organizing the data for DESeq2
RNA-Seq with Bioconductor in R

Mary Piper
Bioinformatics Consultant and Trainer
Bringing in data for DESeq2: sample order
Metadata
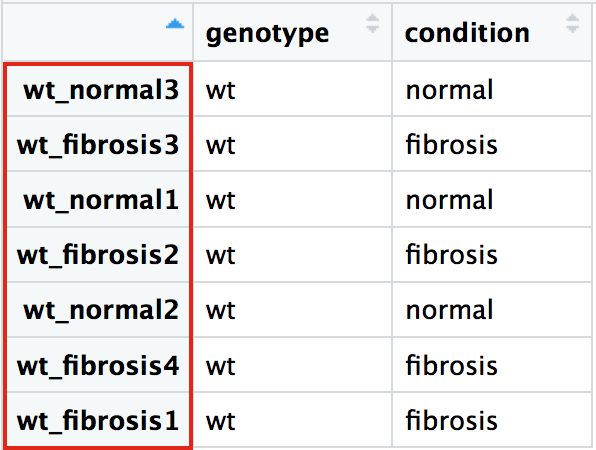
Raw counts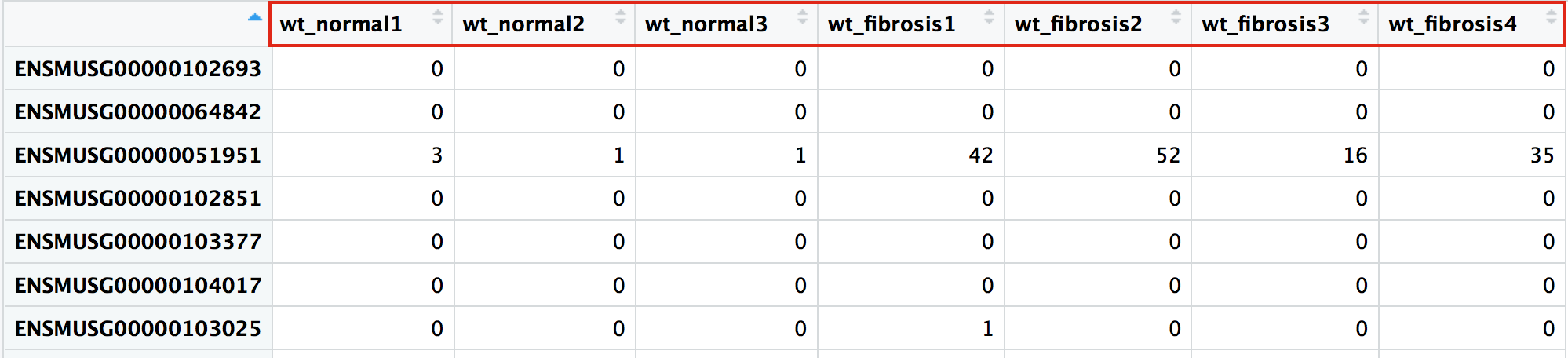
Bringing in data for DESeq2: sample order
rownames(wt_metadata)
[1] "wt_normal3" "smoc2_fibrosis2" "wt_fibrosis3" "smoc2_fibrosis3" "smoc2_normal3" "wt_normal1"
[7] "smoc2_normal4" "wt_fibrosis2" "wt_normal2" "smoc2_normal1" "smoc2_fibrosis1" "smoc2_fibrosis4"
[13] "wt_fibrosis4" "wt_fibrosis1"
colnames(wt_rawcounts)
[1] "wt_normal1" "wt_normal2" "wt_normal3" "wt_fibrosis1" "wt_fibrosis2" "wt_fibrosis3"
[7] "wt_fibrosis4" "smoc2_normal1" "smoc2_normal3" "smoc2_normal4" "smoc2_fibrosis1" "smoc2_fibrosis2"
[13] "smoc2_fibrosis3" "smoc2_fibrosis4"
Bringing in data for DESeq2: sample order
all(rownames(wt_metadata) == colnames(wt_rawcounts))
FALSE
Matching order between vectors
Using the match() function:
match(vector1, vector2)
vector1: vector of values with the desired order
vector2: vector of values to reorder
output: the indices for how to rearrange vector2 to be in the same order as vector1
match(colnames(wt_rawcounts), rownames(wt_metadata)
6 9 1 14 8 3 13 10 5 7 11 2 4 12
Reordering using match() output:
idx <- match(colnames(wt_rawcounts), rownames(wt_metadata))reordered_wt_metadata <- wt_metadata[idx, ]View(reordered_wt_metadata)
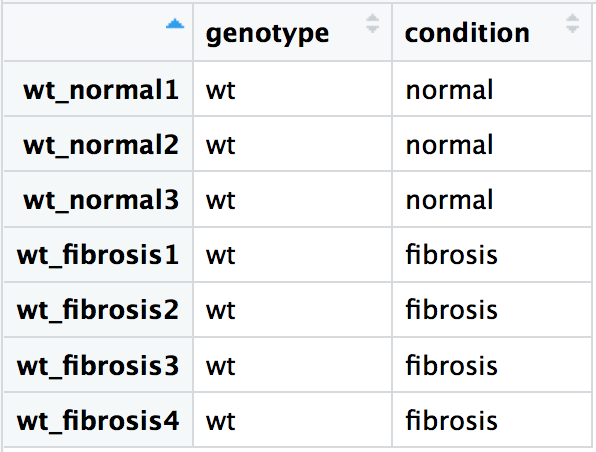
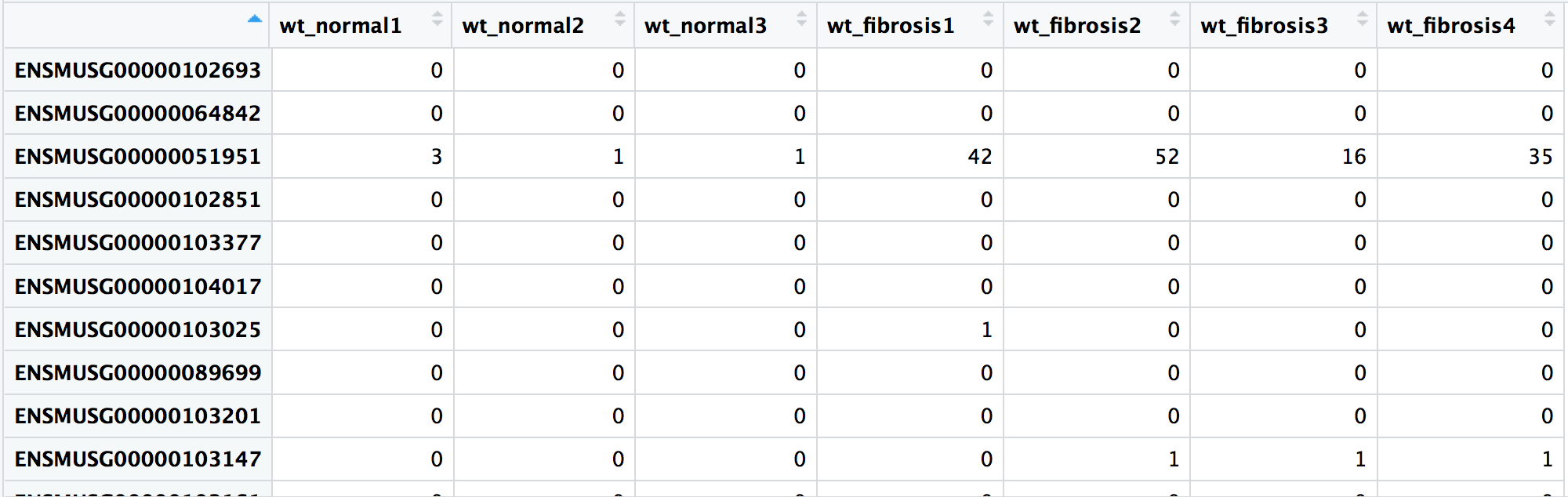
all(rownames(reordered_wt_metadata) == colnames(wt_rawcounts))
TRUE

Creating the DESeq2 object
# Create DESeq object
dds_wt <- DESeqDataSetFromMatrix(countData = wt_rawcounts,
colData = reordered_wt_metadata,
design = ~ condition)
Let's practice!
RNA-Seq with Bioconductor in R

