DESeq2 results
RNA-Seq with Bioconductor in R

Mary Piper
Bioinformatics Consultant and Trainer
DESeq2 results table
mcols(wt_res)

head(wt_res, n=10)
log2 fold change (MAP): condition fibrosis vs normal
Wald test p-value: condition fibrosis vs normal
data frame with 6 rows and 6 columns
baseMean log2FoldChange lfcSE stat
<numeric> <numeric> <numeric> <numeric>
ENSMUSG00000102693 0 NA NA NA
ENSMUSG00000064842 0 NA NA NA
ENSMUSG00000051951 19.5084656230804 3.55089043143673 0.648400500074659 4.66871842838828
ENSMUSG00000102851 0 NA NA NA
ENSMUSG00000103377 0 NA NA NA
ENSMUSG00000104017 0 NA NA NA
pvalue padj
<numeric> <numeric>
ENSMUSG00000102693 NA NA
ENSMUSG00000064842 NA NA
ENSMUSG00000051951 3.03084428526558e-06 1.93776447202312e-05
ENSMUSG00000102851 NA NA
ENSMUSG00000103377 NA NA
ENSMUSG00000104017 NA NA
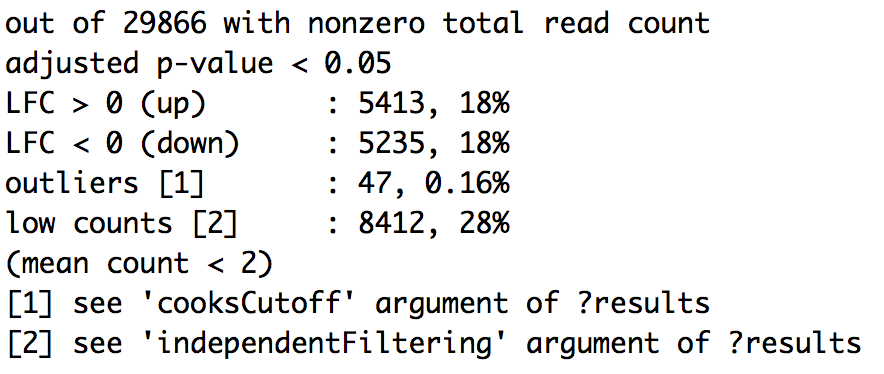
Significant DE genes - summary
summary(wt_res)
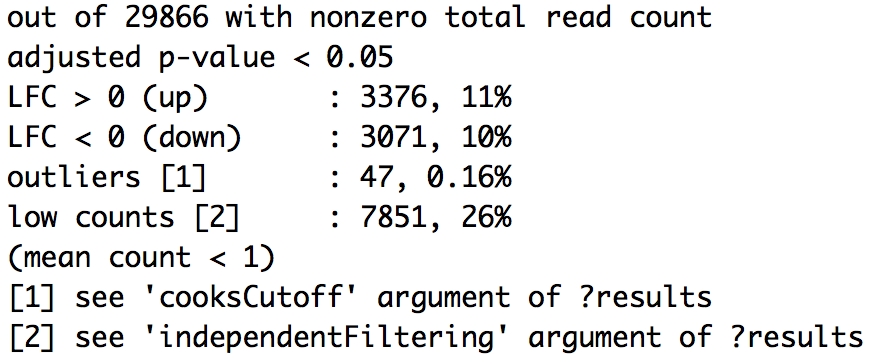
Significant DE genes - fold-change threshold
wt_res <- results(dds_wt,
contrast = c("condition", "fibrosis", "normal"),
alpha = 0.05,
lfcThreshold = 0.32)
wt_res <- lfcShrink(dds_wt,
contrast=c("condition", "fibrosis", "normal"),
res=wt_res)
Significant DE genes - summary
summary(wt_res)
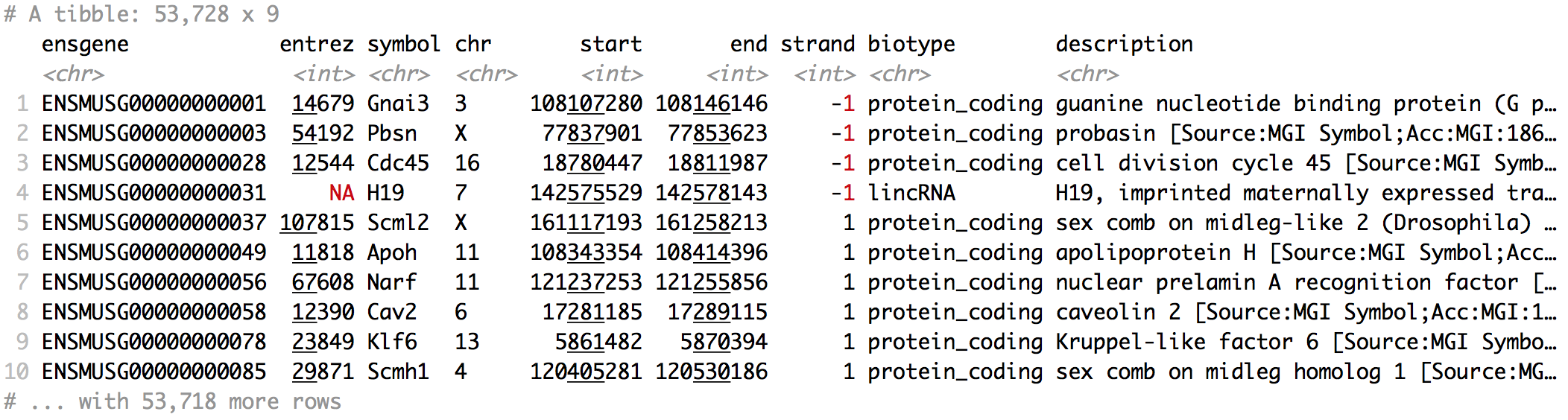
Results - annotate
library(annotables)
grcm38
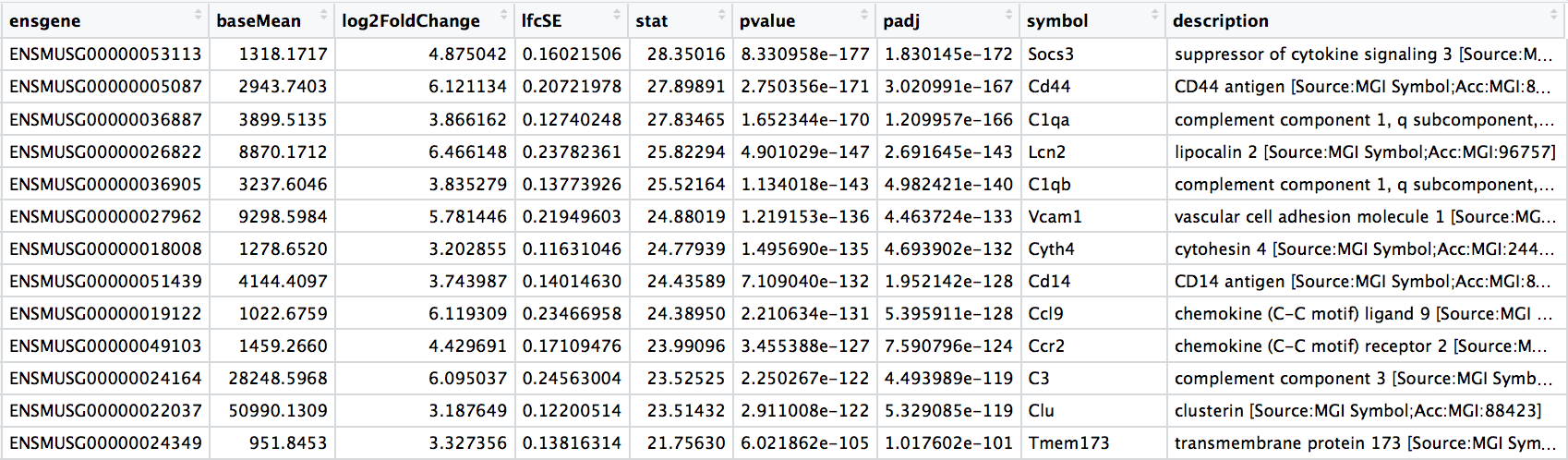
Results - extract
wt_res_all <- data.frame(wt_res) %>%
rownames_to_column(var = "ensgene") %>%
left_join(x = wt_res_all,
y = grcm38[, c("ensgene", "symbol", "description")],
by = "ensgene")
View(wt_res_all)
Significant DE genes - arrange
wt_res_sig <- subset(wt_res_all, padj < 0.05)wt_res_sig <- wt_res_sig %>% arrange(padj)View(wt_res_all)


Let's practice!
RNA-Seq with Bioconductor in R

