Differential gene expression overview
RNA-Seq with Bioconductor in R

Mary Piper
Bioinformatics Consultant and Trainer

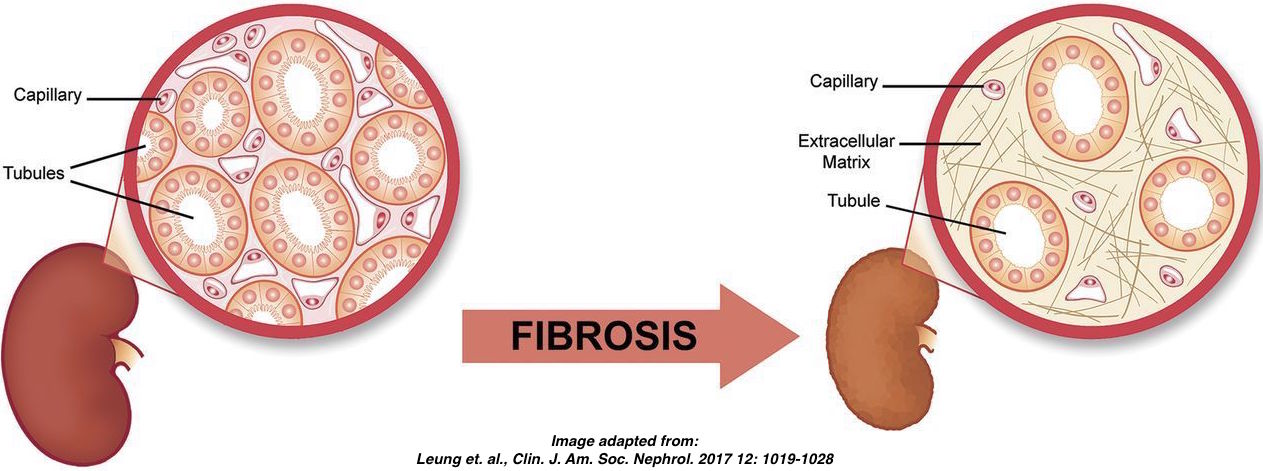
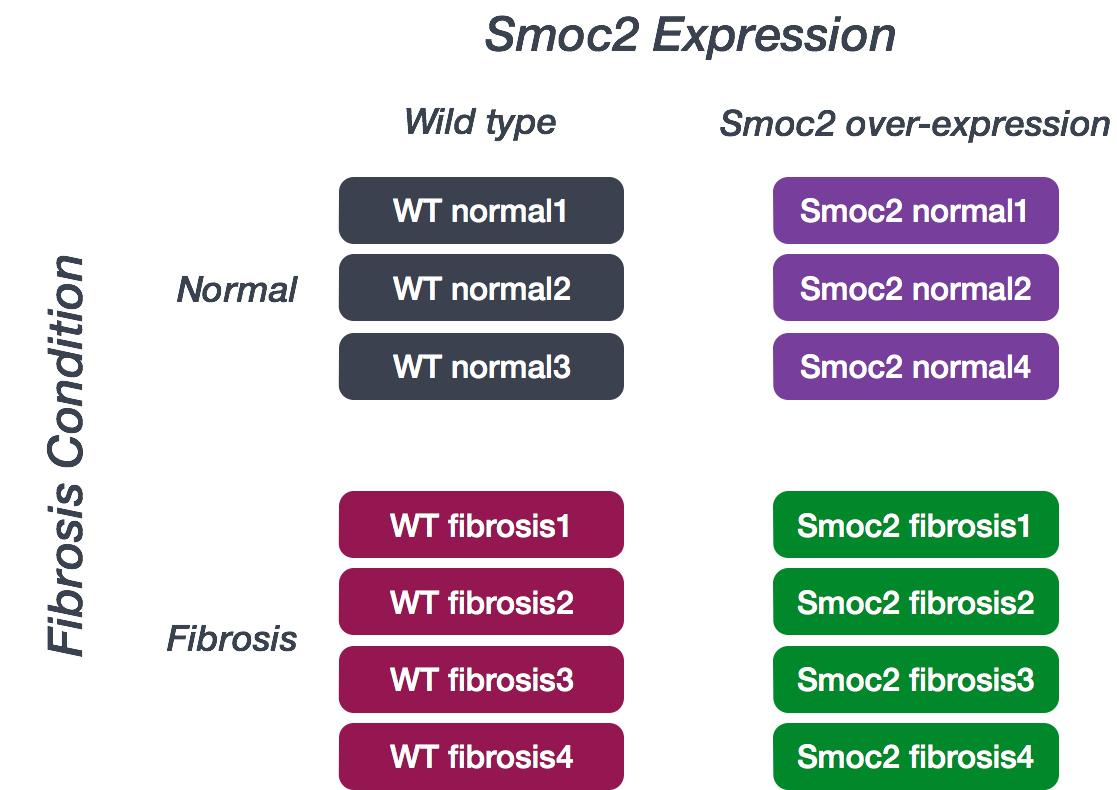
Introduction to dataset: Smoc2
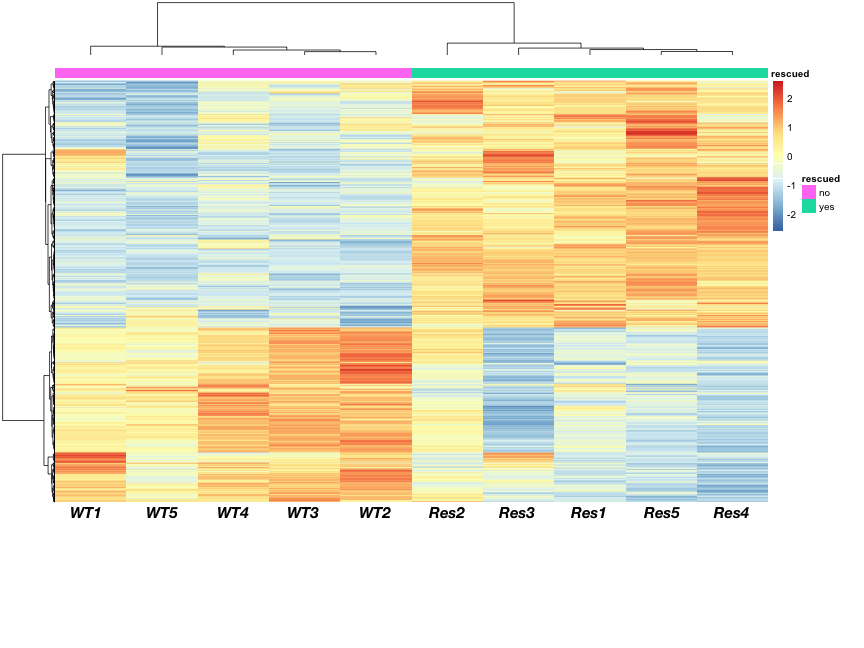
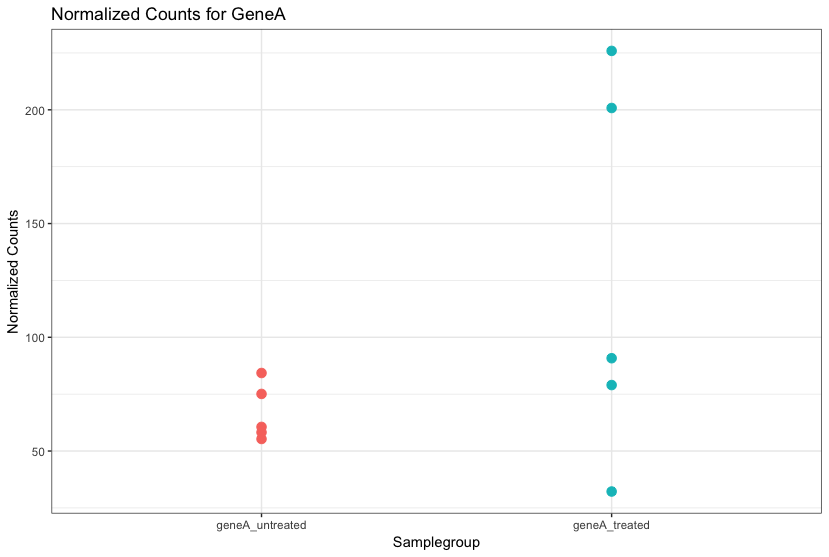
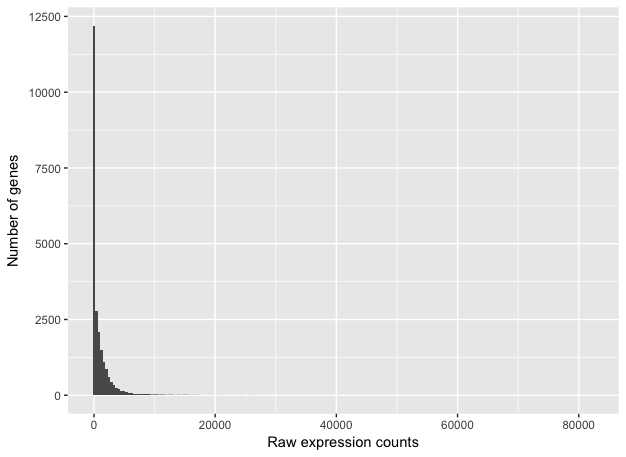
RNA-Seq count distribution
ggplot(raw_counts) +
geom_histogram(aes(x = wt_normal1), stat = "bin", bins = 200) +
xlab("Raw expression counts") +
ylab("Number of genes")
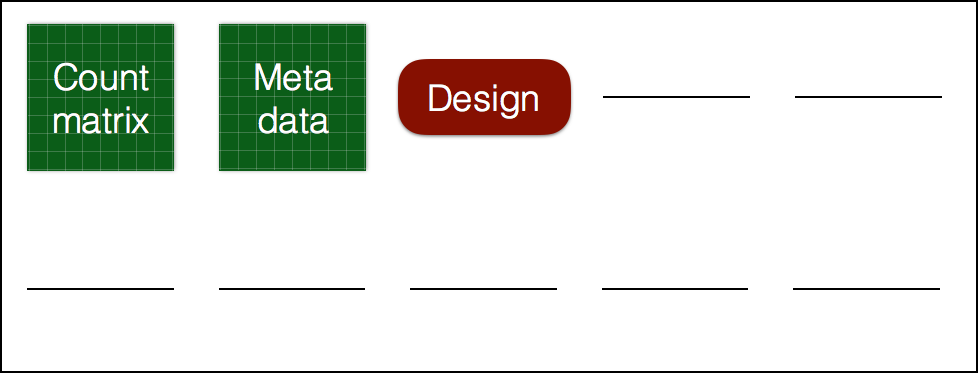
Preparation for differential expression analysis: DESeq2 object
dds <- DESeqDataSetFromMatrix(countData = rawcounts,
colData = metadata,
design = ~ condition)
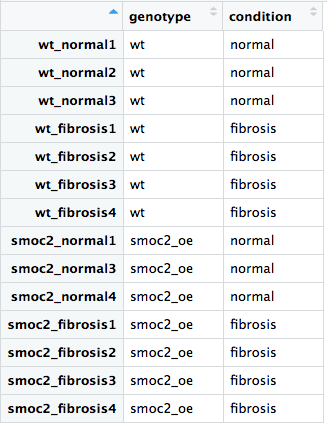
Preparation for differential expression analysis: metadata
# Create vectors containing metadata for the samples genotype <- c("wt", "wt", "wt", "wt", "wt", "wt", "wt") condition <- c("normal", "fibrosis", "normal", "fibrosis", "normal", "fibrosis", "fibrosis") # Combine vectors into a data frame wt_metadata <- data.frame(genotype, wildtype)# Create the row names with the associated sample names rownames(wt_metadata) <- c("wt_normal3", "wt_fibrosis3", "wt_normal1", "wt_fibrosis2", "wt_normal2", "wt_fibrosis4", "wt_fibrosis1")
Preparation for differential expression analysis: metadata
Let's practice!
RNA-Seq with Bioconductor in R

