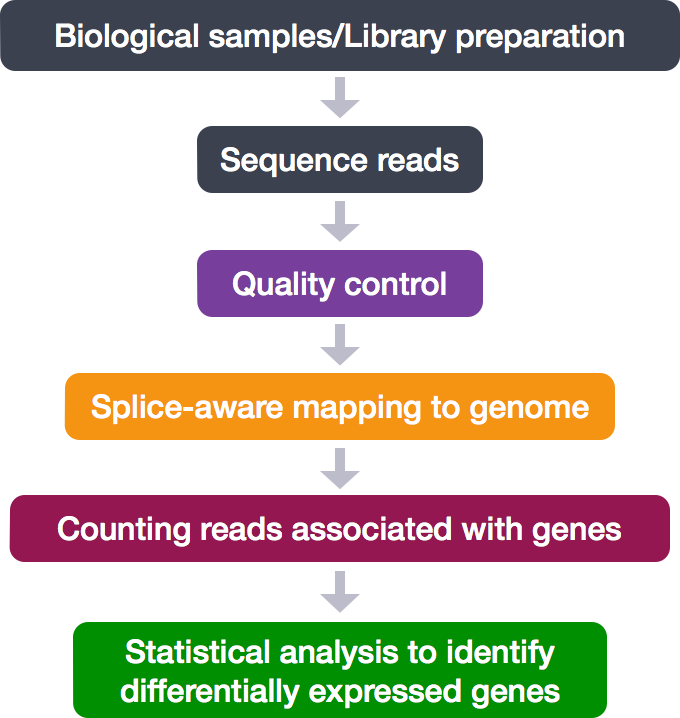
RNA-Seq Workflow
RNA-Seq with Bioconductor in R

Mary Piper
Bioinformatics Consultant and Trainer
RNA-Seq Workflow: RNA-Seq Experimental Design
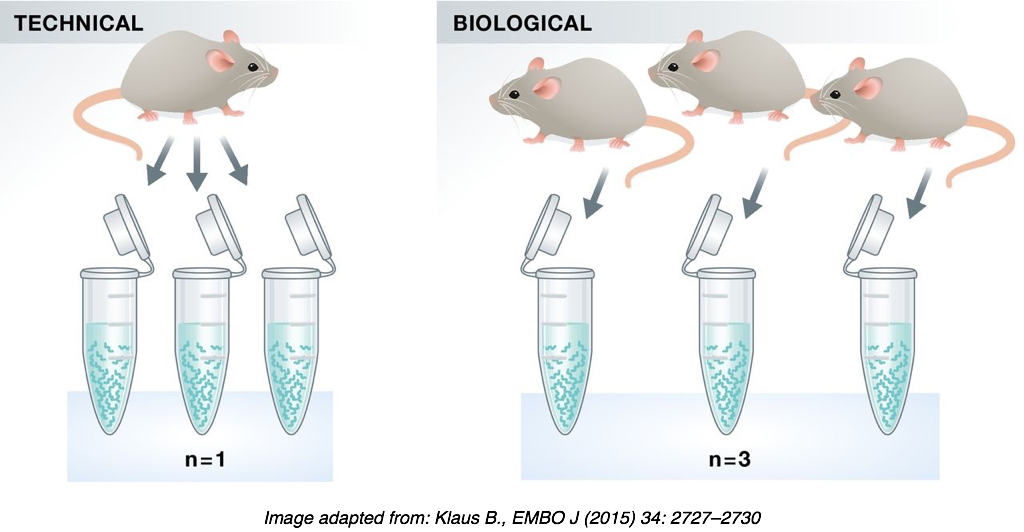
Technical replicates: Generally low technical variation, so unnecessary.
Biological replicates: Crucial to the success of RNA-Seq differential expression analyses. The more replicates the better, but at the very least have 3.
Batch effects: Avoid as much as possible and note down all experimental variables.
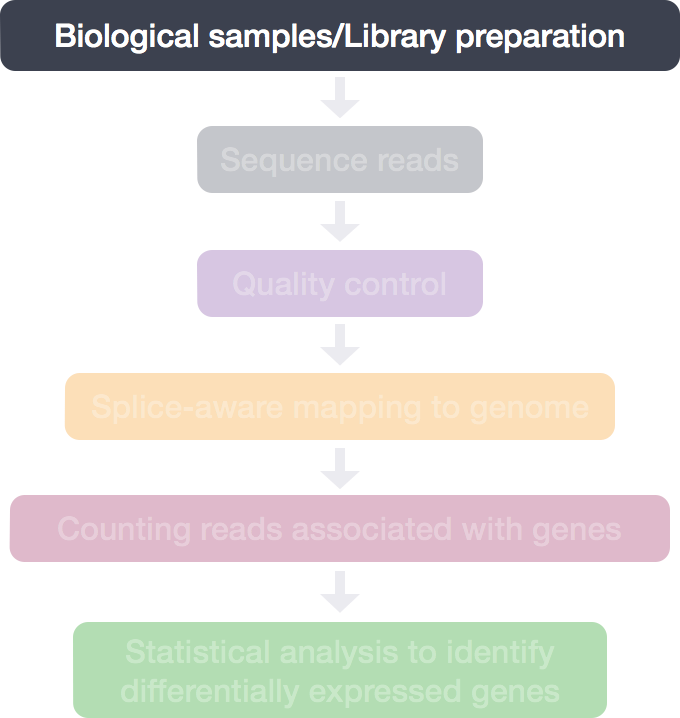
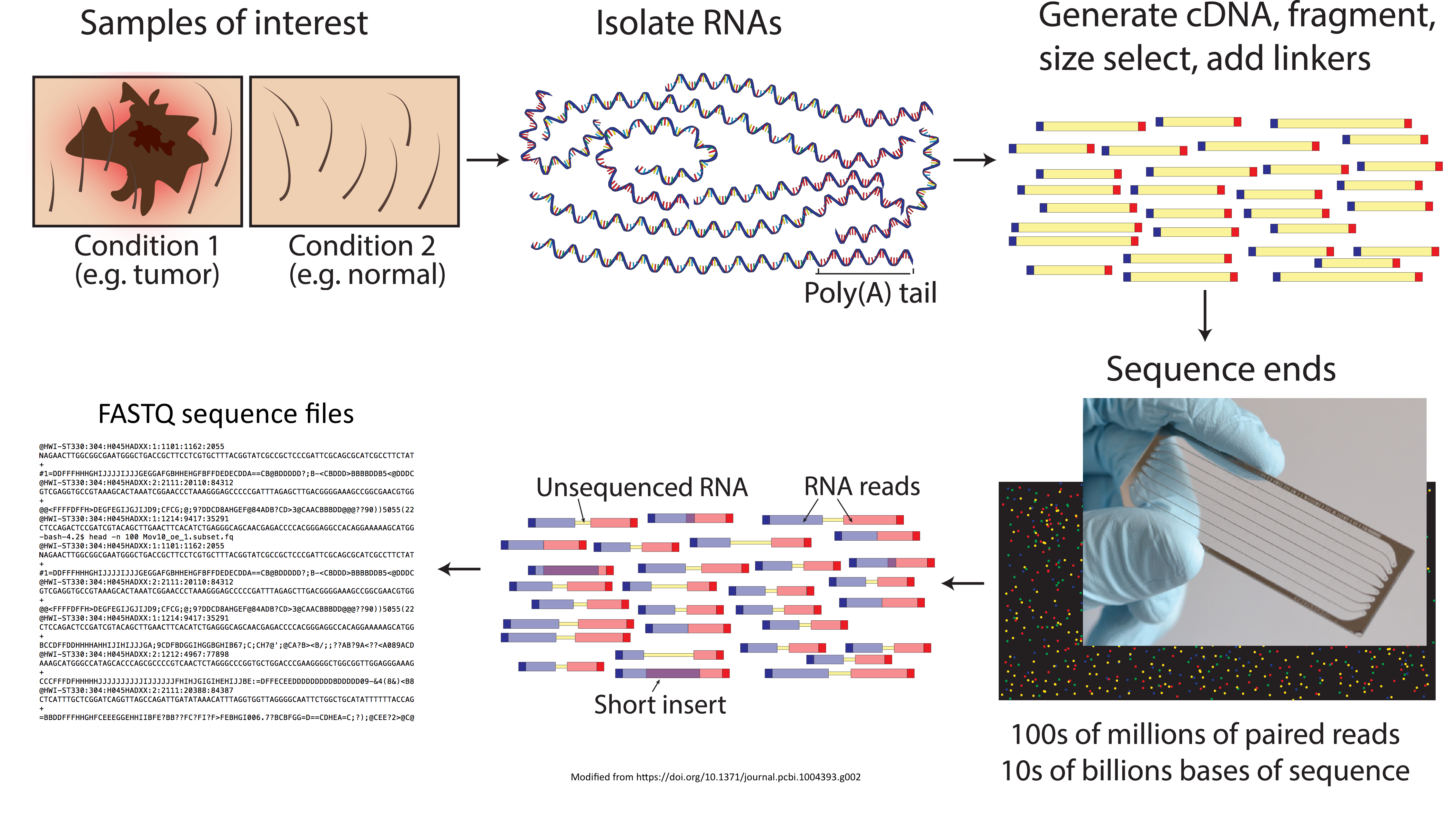
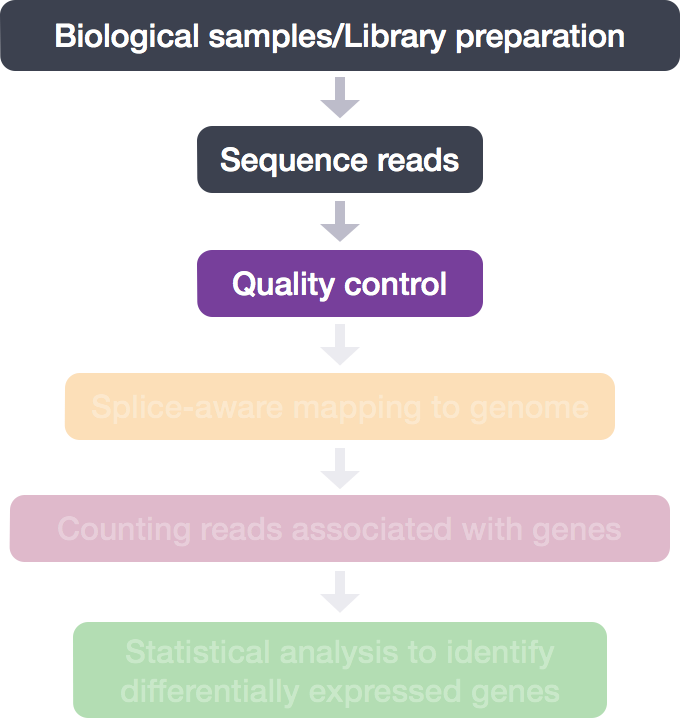
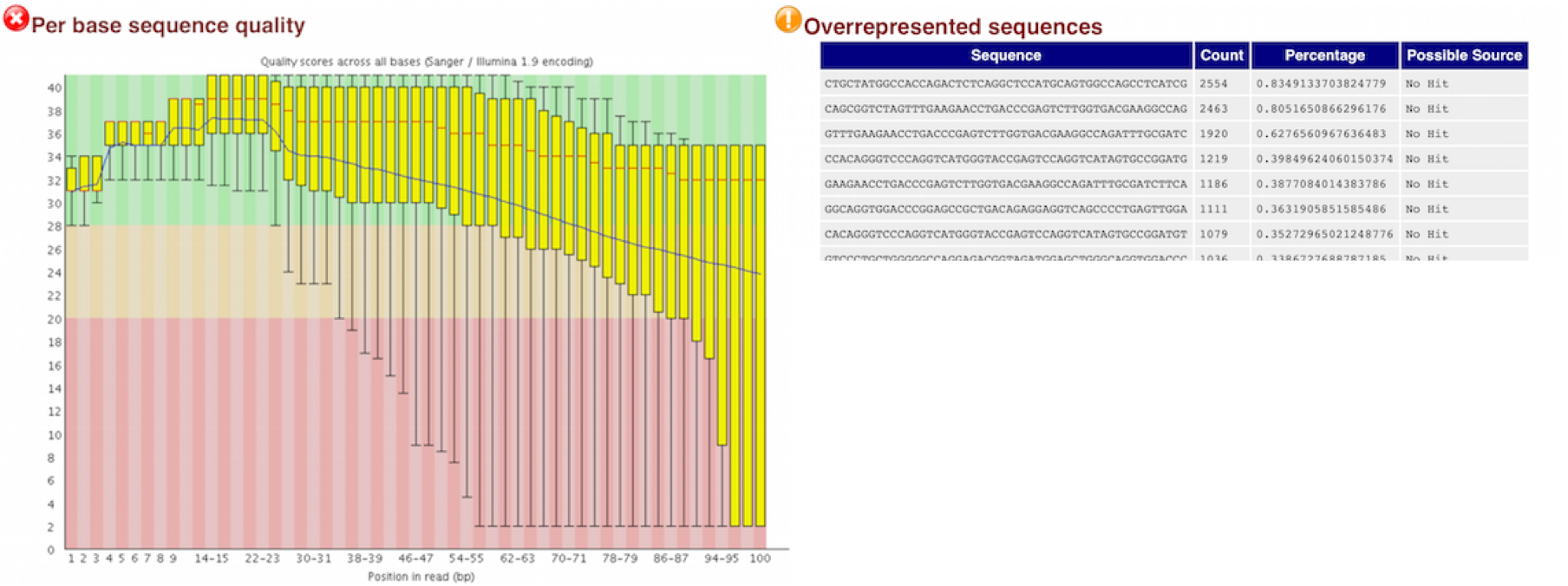
RNA-Seq Workflow: Quality control
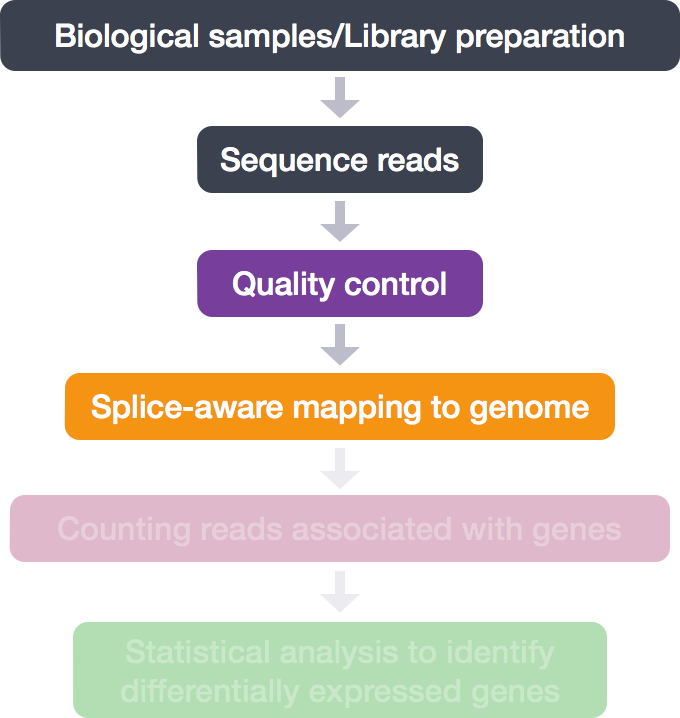
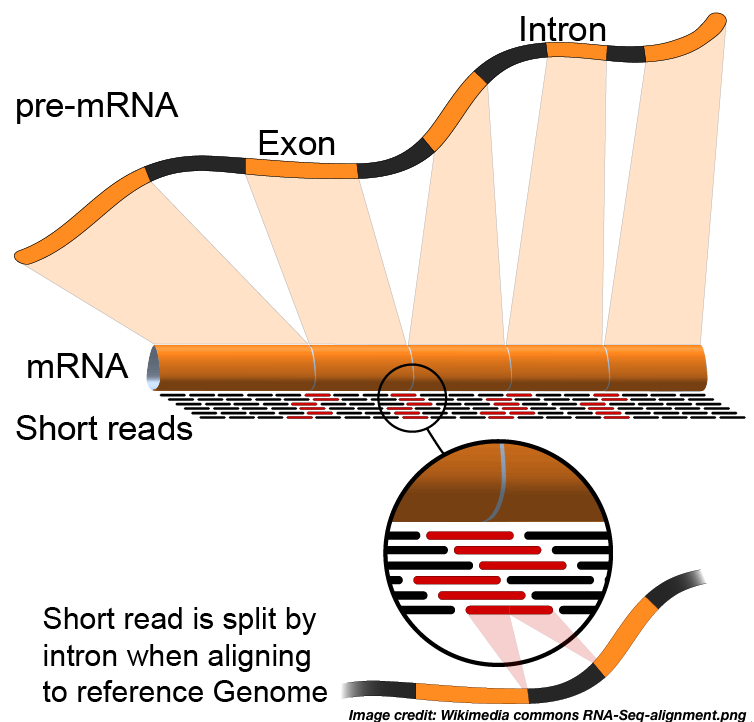
RNA-Seq Workflow: Alignment
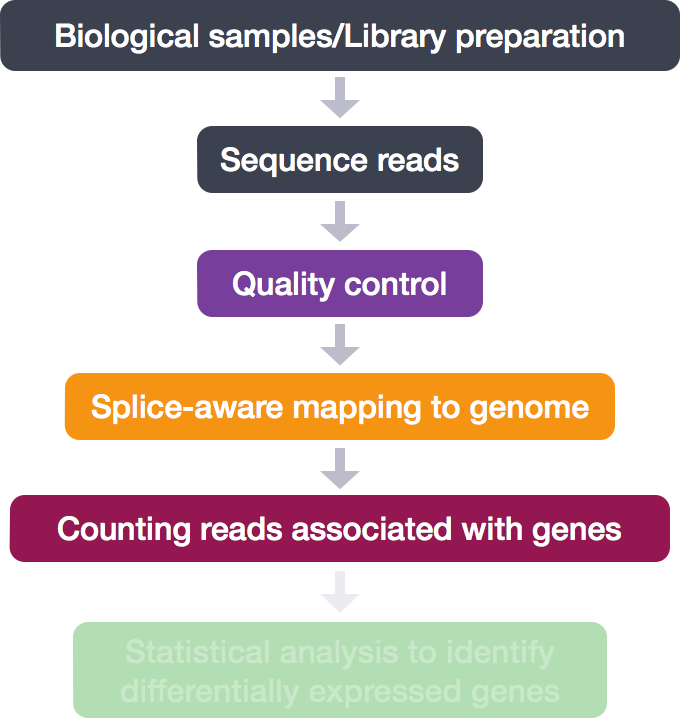
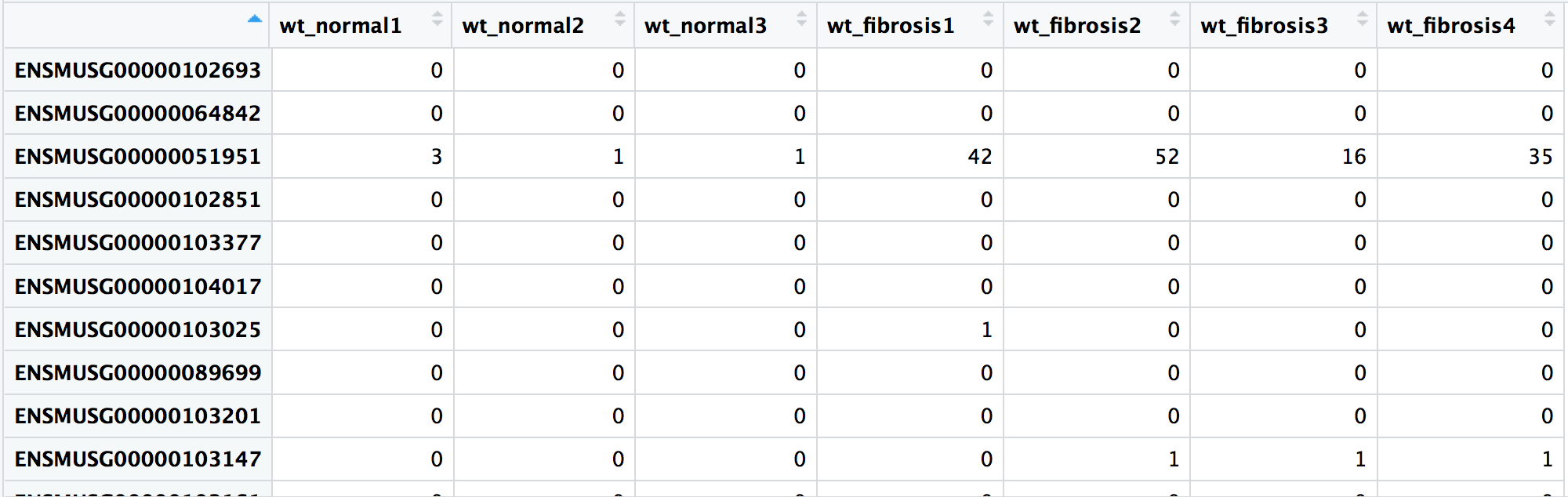
RNA-Seq Workflow: Count matrix
wt_rawcounts <- read.csv("fibrosis_wt_rawcounts.csv")
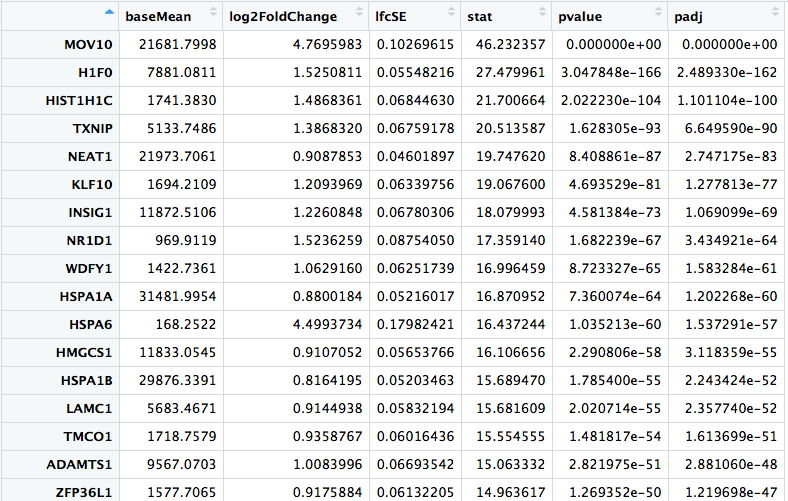

Back to you!
RNA-Seq with Bioconductor in R

