Visualizing the Cox model
Survival Analysis in R

Heidi Seibold
Statistician at LMU Munich
Steps to visualize a Cox model
- Compute Cox model
- Decide on covariate combinations ("imaginary patients")
- Compute survival curves
- Create
data.framewith survival curve information - Plot
Step 1: Compute Cox model
cxmod <- coxph(Surv(time, cens) ~ horTh + tsize, data = GBSG2)
- Decide on covariate combinations ("imaginary patients")
newdat <- expand.grid( horTh = levels(GBSG2$horTh), tsize = quantile(GBSG2$tsize, probs = c(0.25, 0.5, 0.75)) ) rownames(newdat) <- letters[1:6] newdat
horTh tsize
a no 20
b yes 20
c no 25
...
Step 2: Compute survival curves
cxsf <- survfit(cxmod, data = GBSG2, newdata = newdat, conf.type = "none")str(cxsf)
List of 10
$ n : int 686
$ time : num [1:574] 8 15 16 17 18 29 42 46 57 63 ...
$ n.risk : num [1:574] 686 685 684 683 681 680 679 678 677 676 ...
$ n.event : num [1:574] 0 0 0 0 0 0 0 0 0 0 ...
$ n.censor: num [1:574] 1 1 1 2 1 1 1 1 1 1 ...
$ surv : num [1:574, 1:6] 1 1 1 1 1 1 1 1 1 1 ...
..- attr(*, "dimnames")=List of 2
.. ..$ : NULL
.. ..$ : chr [1:6] "a" "b" "c" "d" ...
$ type : chr "right"
$ cumhaz : num [1:574, 1:6] 0 0 0 0 0 0 0 0 0 0 ...
$ std.err : num [1:574, 1:6] 0 0 0 0 0 0 0 0 0 0 ...
$ call : language survfit(formula = cxmod, newdata = newdat, conf.type = "none", data = GBSG2)
- attr(*, "class")= chr [1:2] "survfit.cox" "survfit"
Step 3: Create data.frame with survival curve information
surv_cxmod0 <- surv_summary(cxsf)
head(surv_cxmod0)
time n.risk n.event n.censor surv std.err upper lower strata
1 8 686 0 1 1 0 NA NA a
2 15 685 0 1 1 0 NA NA a
3 16 684 0 1 1 0 NA NA a
4 17 683 0 2 1 0 NA NA a
5 18 681 0 1 1 0 NA NA a
6 29 680 0 1 1 0 NA NA a
surv_cxmod <- cbind(surv_cxmod0,
newdat[as.character(surv_cxmod0$strata), ])
Step 4: Plot
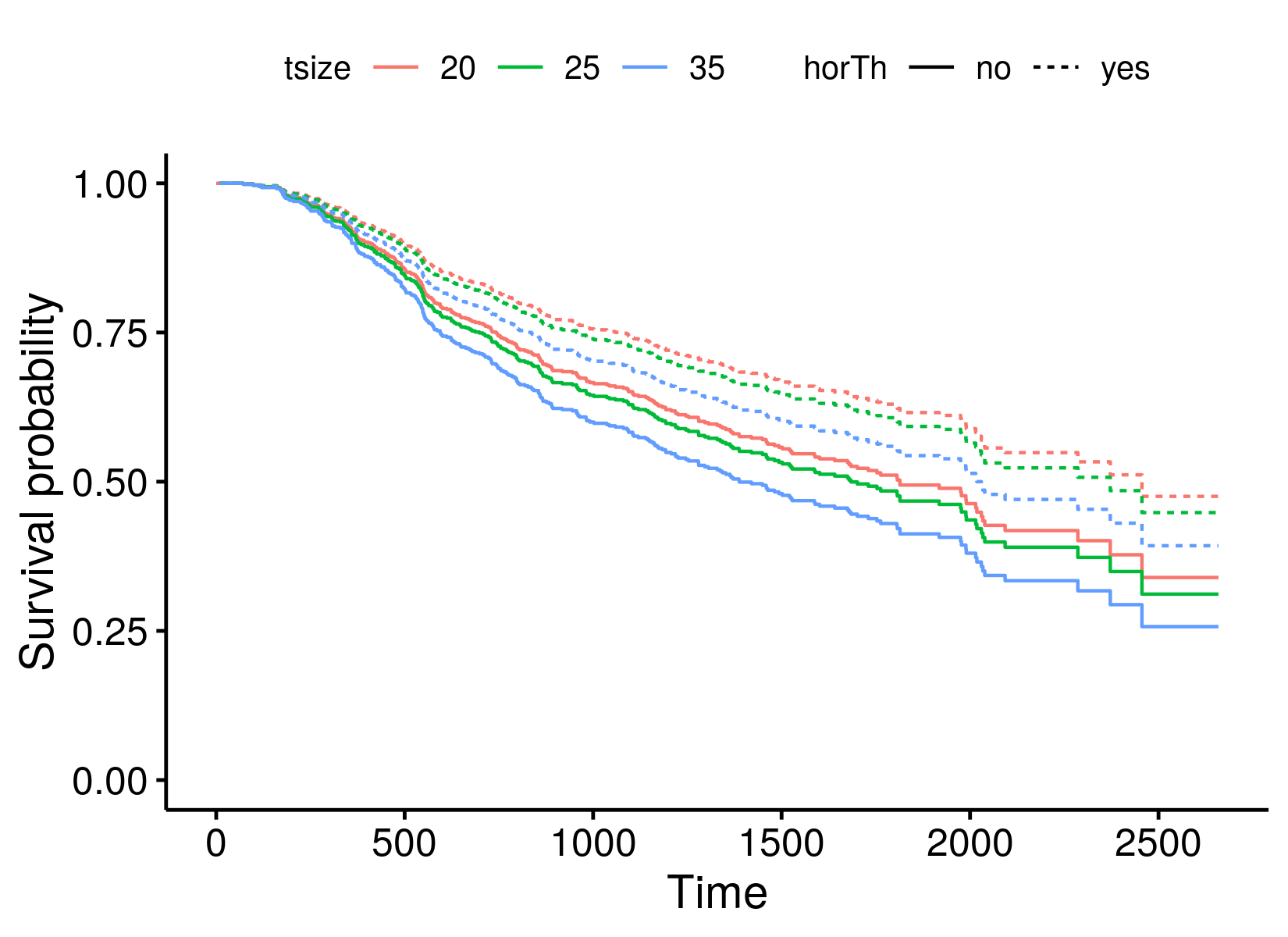
ggsurvplot_df(surv_cxmod, linetype = "horTh", color = "tsize",
legend.title = NULL, censor = FALSE)
Now it's your turn to visualize!
Survival Analysis in R

