Visualising Weibull models
Survival Analysis in R

Heidi Seibold
Statistician at LMU Munich
Steps to produce visualization
- Compute Weibull model
- Decide on "imaginary patients"
- Compute survival curves
- Create
data.framewith survival curve information - Plot

Step 1 and 2
- Compute Weibull model
wbmod <- survreg(Surv(time, cens) ~ horTh + tsize, data = GBSG2) - Decide on Decide on covariate combinations ("imaginary patients")
newdat <- expand.grid( horTh = levels(GBSG2$horTh), tsize = quantile(GBSG2$tsize, probs = c(0.25, 0.5, 0.75)) ) newdat
#> horTh tsize
#> 1 no 20
#> 2 yes 20
#> 3 no 25
...
Step 3
- Compute survival curves
surv <- seq(.99, .01, by = -.01) t <- predict(wbmod, type = "quantile", p = 1 - surv, newdata = newdat)dim(t) t[, 1:7]
6 99
[,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 65.86524 112.54061 154.2116 193.0603 230.0268 265.6298 300.1952
[2,] 89.96016 153.71037 210.6256 263.6858 314.1755 362.8029 410.0131
[3,] 61.97352 105.89102 145.0999 181.6531 216.4354 249.9348 282.4579
[4,] 84.64477 144.62823 198.1805 248.1057 295.6121 341.3663 385.7870
[5,] 54.86634 93.74733 128.4597 160.8209 191.6144 221.2720 250.0653
[6,] 74.93762 128.04211 175.4530 219.6526 261.7110 302.2180 341.5445
Step 4
- Create
data.framewith survival curve information
surv_wbmod_wide <- cbind(newdat, t)
library("reshape2")
surv_wbmod <- melt(surv_wbmod_wide, id.vars = c("horTh", "tsize"),
variable.name = "surv_id", value.name = "time")
surv_wbmod$surv <- surv[as.numeric(surv_wbmod$surv_id)]
surv_wbmod[, c("upper", "lower", "std.err", "strata")] <- NA
Step 4
str(surv_wbmod)
'data.frame': 594 obs. of 9 variables:
$ horTh : Factor w/ 2 levels "no","yes": 1 2 1 2 1 2 1 2 1 2 ...
$ tsize : num 20 20 25 25 35 35 20 20 25 25 ...
$ surv_id: Factor w/ 99 levels "1","2","3","4",..: 1 1 1 1 1 1 2 2 2 2 ...
$ time : num 65.9 90 62 84.6 54.9 ...
$ surv : num 0.99 0.99 0.99 0.99 0.99 0.99 0.98 0.98 0.98 0.98 ...
$ strata : logi NA NA NA NA NA NA ...
$ std.err: logi NA NA NA NA NA NA ...
$ lower : logi NA NA NA NA NA NA ...
$ upper : logi NA NA NA NA NA NA ...
If this was too fast...
library("reshape2")
surv_wbmod <- melt(surv_wbmod_wide, id.vars = names(newdat),
variable.name = "surv_id", value.name = "time")
?melt
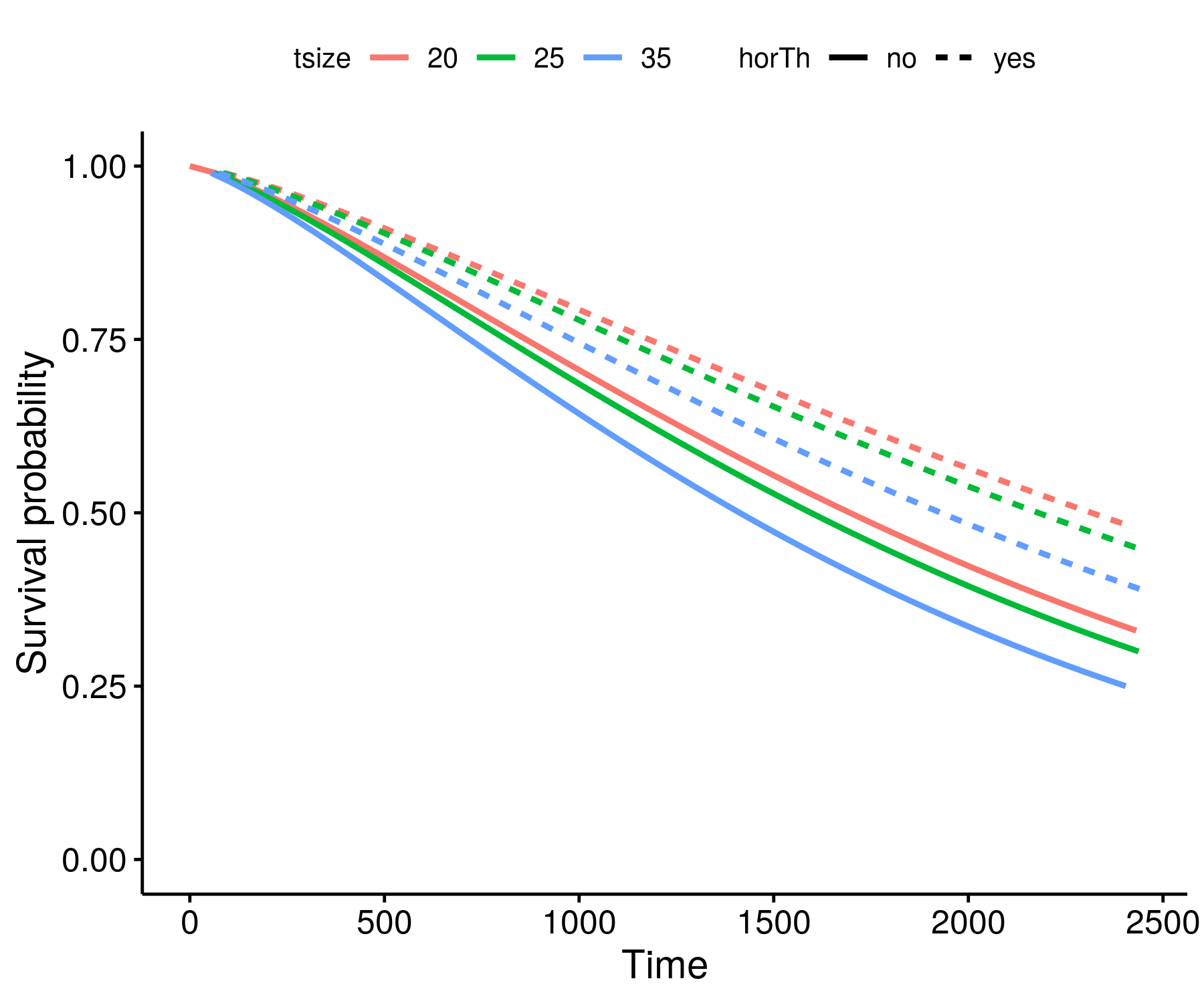
Step 5
- Plot
ggsurvplot_df(surv_wbmod, surv.geom = geom_line,
linetype = "horTh", color = "tsize", legend.title = NULL)
You can do it too! All you need is practice.
Survival Analysis in R

