Measuring Intensity
Biomedical Image Analysis in Python

Stephen Bailey
Instructor
Measuring intensity
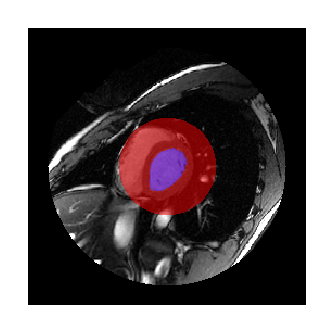
We have the following labels for a single volume of the cardiac time series:
- Left ventricle
- Central portion
Functions
scipy.ndimage.measurements
ndi.mean()
ndi.median()
ndi.sum()
ndi.maximum()
ndi.standard_deviation()
ndi.variance()
Functions applied over all dimensions, optionally at specific labels.
Custom functions:
ndi.labeled_comprehension()
Calling measurement functions
import imageio import scipy.ndimage as ndi vol=imageio.volread('SCD-3d.npz') label=imageio.volread('labels.npz')# All pixels ndi.mean(vol)
3.7892
# Labeled pixels
ndi.mean(vol, label)
89.2342
# Label 1
ndi.mean(vol, label, index=1)
163.2930
# Labels 1 and 2
ndi.mean(vol, label, index=[1,2])
[163.2930, 60.2847]
Object histograms
hist=ndi.histogram(vol, min=0, max=255, bins=256)obj_hists=ndi.histogram(vol, 0, 255, 256, labels, index=[1, 2]) len(obj_hists)
2
Object histograms
plt.plot(obj_hists[0],
label='Left ventricle')
plt.plot(obj_hists[1],
label='Other labelled pixels')
plt.legend()
plt.show()
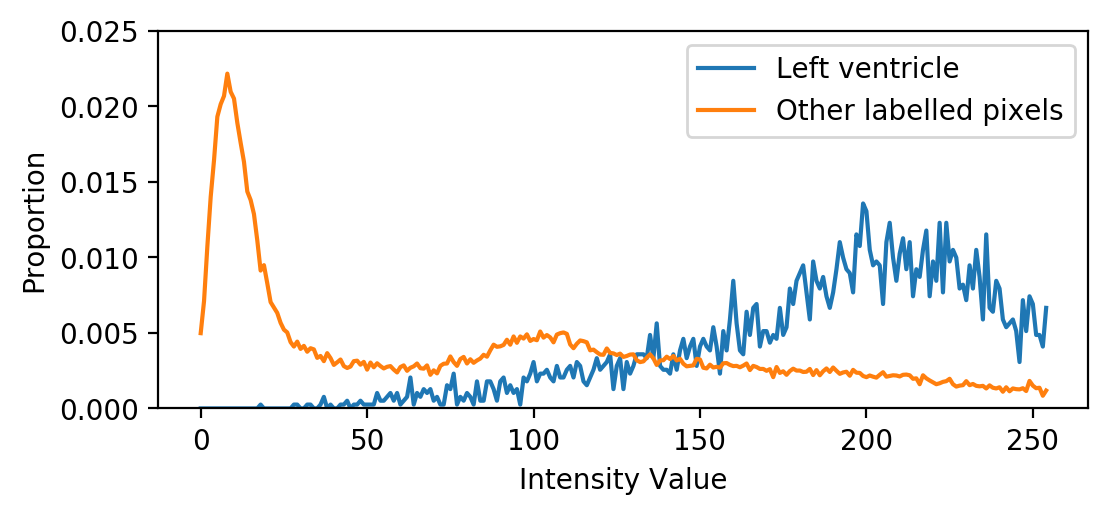
Histograms containing multiple tissue types will have several peaks
Histograms for well-segmented tissue often resemble a normal distribution
Let's practice!
Biomedical Image Analysis in Python

