Spatial transformations
Biomedical Image Analysis in Python

Stephen Bailey
Instructor
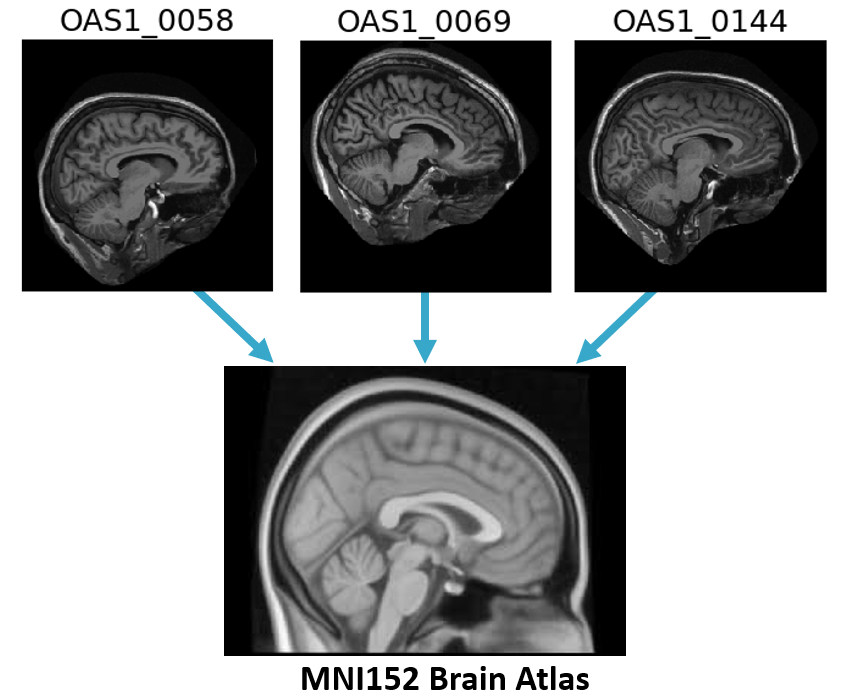
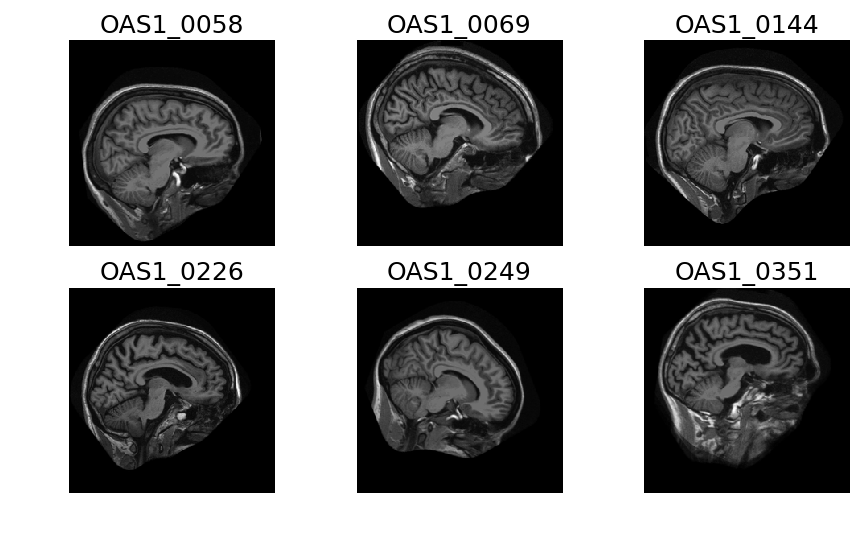
OASIS Database
Significant variability
Registration
- Align images to template
Minimize spatial variability
Templates:
- may represent multiple subjects
- may be an "average" image
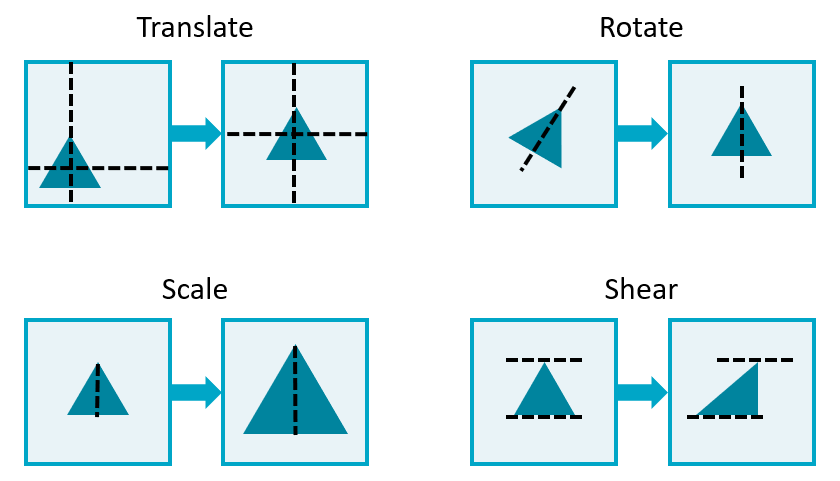
Entails many spatial transformations
Affine transformations preserve points, lines, and planes
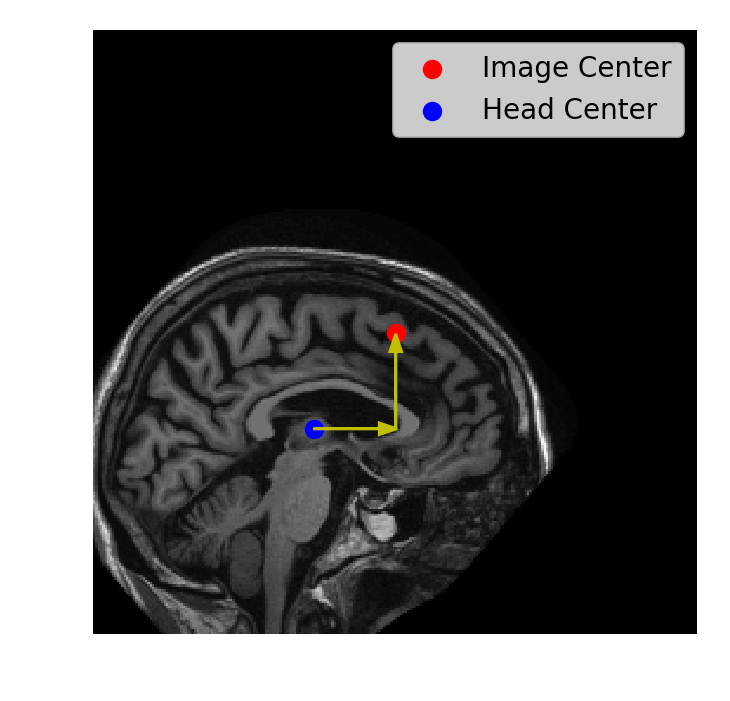
Translation
import imageio
import scipy.ndimage as ndi
im=imageio.imread('OAS1036-2d.dcm')
im.shape
(256, 256)
com = ndi.center_of_mass(im)d0 = 128 - com[0] d1 = 128 - com[1]xfm = ndi.shift(im, shift=[d0, d1])
Rotation
ndi.rotate(im,
angle=25,
axes=(0,1))
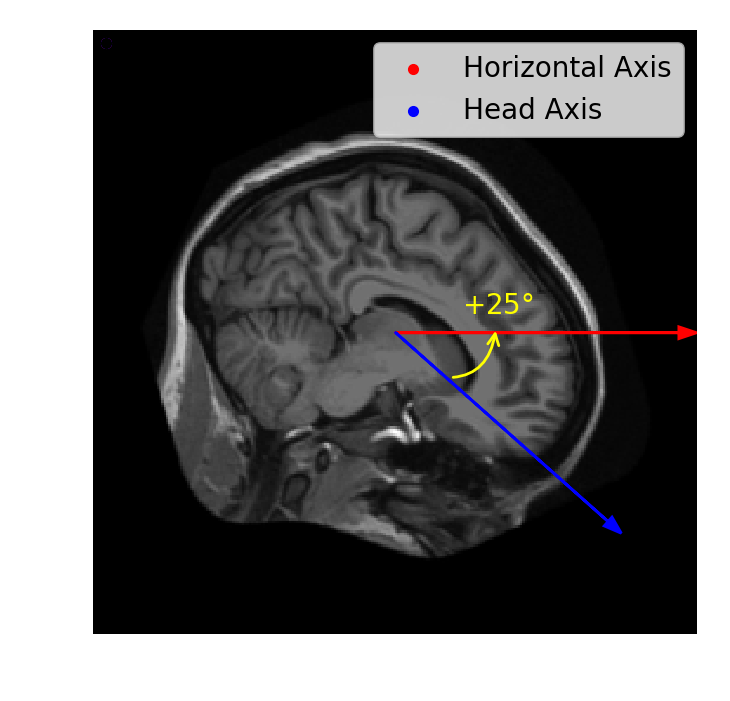
Image rotation
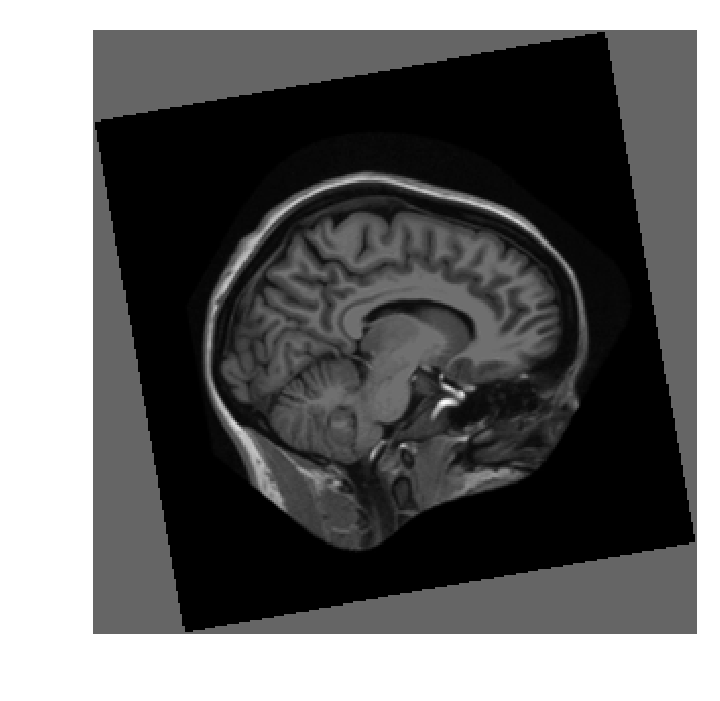
xfm = ndi.rotate(im, angle=25)
xfm.shape
(297, 297)
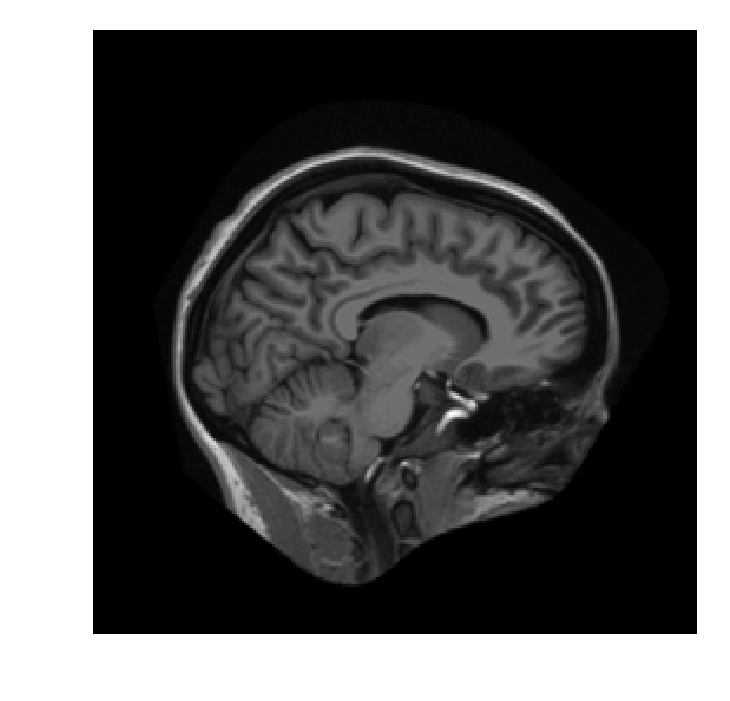
xfm = ndi.rotate(im, angle=25,
reshape=False)
xfm.shape
(256, 256)
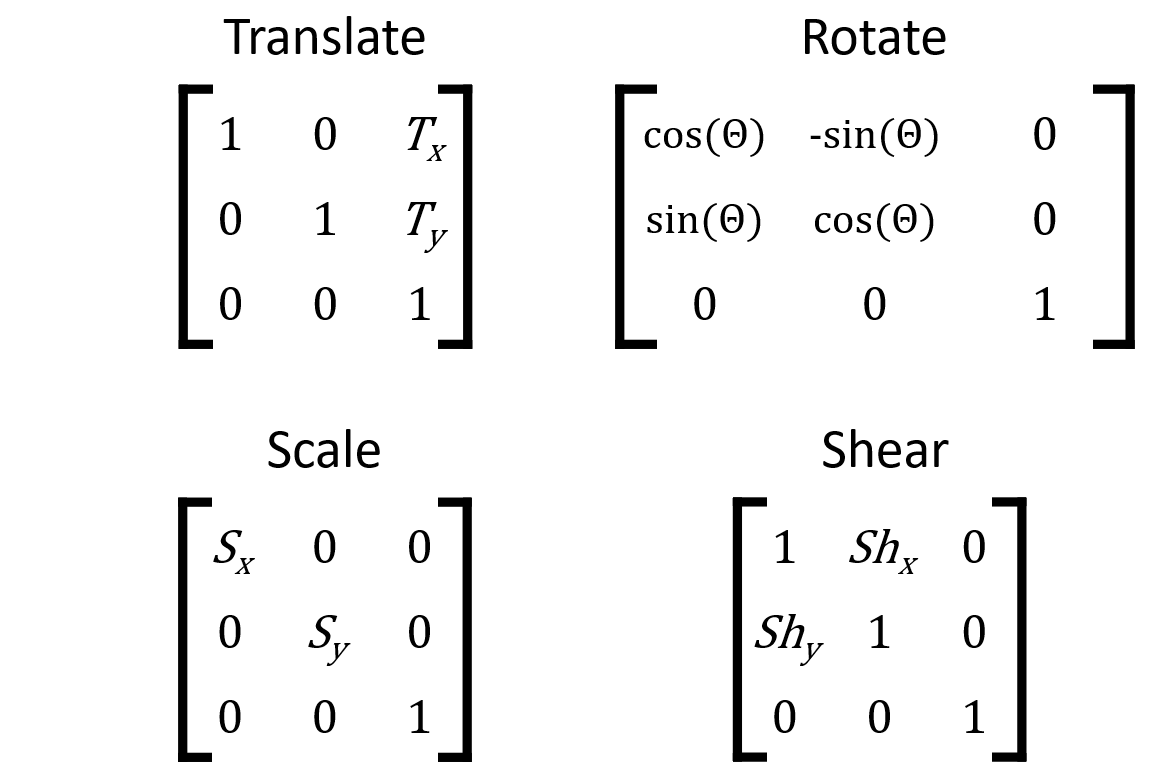
Transformation matrix
Transformation matrix: applied to one image for registration.
Elements of the matrix encode "instructions" for different affine transformations.
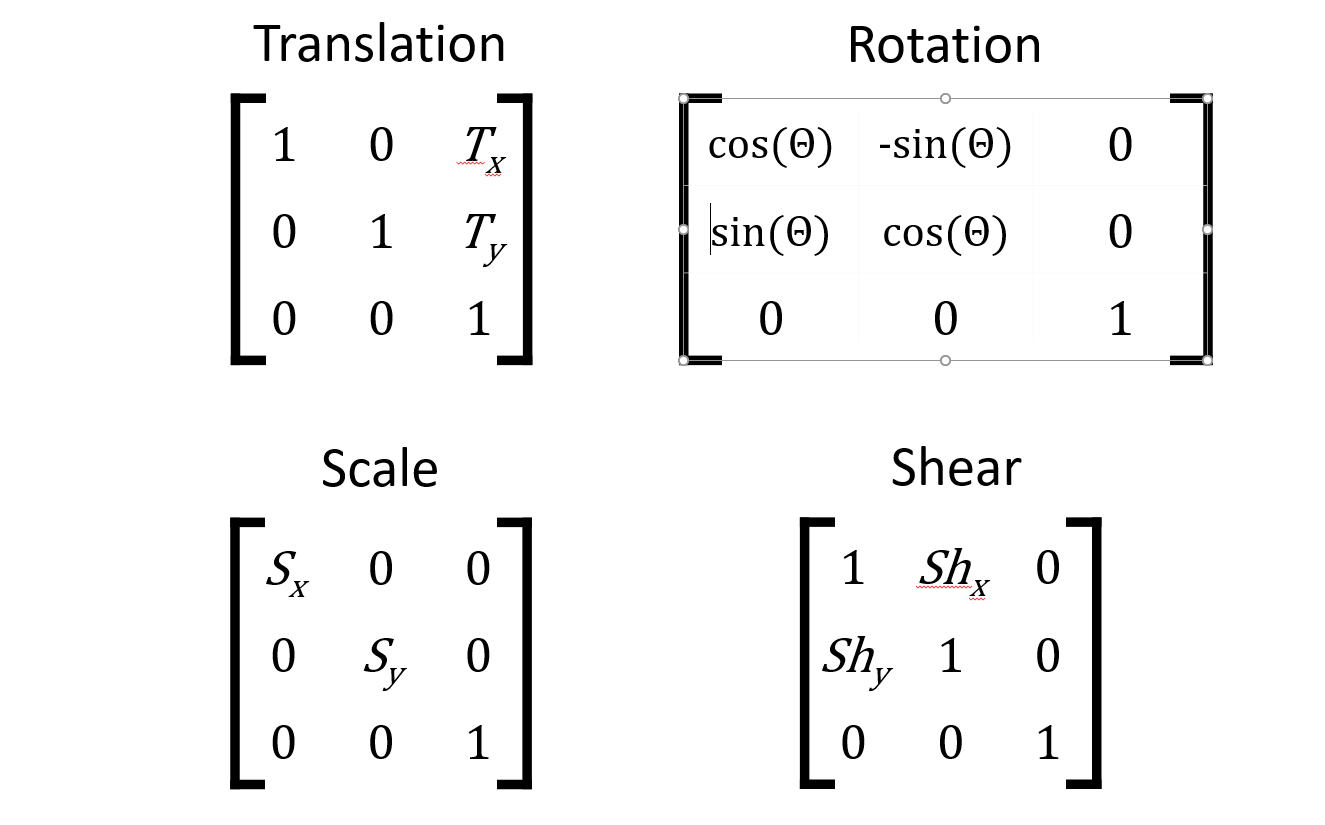
Applying a transformation matrix
# Identity matrix mat = [[1, 0, 0], [0, 1, 0], [0, 0, 1]]xfm = ndi.affine_transform(im, mat)
# Translate and rescale
mat = [[0.8, 0, -20],
[0, 0.8, -10],
[0, 0, 1]]
xfm = ndi.affine_transform(im,
mat)
Let's practice!
Biomedical Image Analysis in Python