Training, test and validation splits
Machine Learning in the Tidyverse

Dmitriy (Dima) Gorenshteyn
Lead Data Scientist, Memorial Sloan Kettering Cancer Center
Train-Test Split
Train-Test Split
Train-Test Split
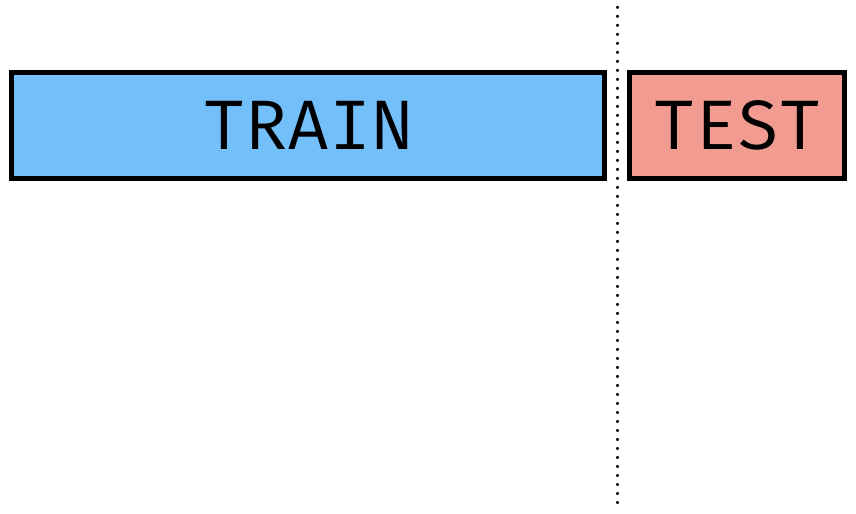
initial_split()
library(rsample) gap_split <- initial_split(gapminder, prop = 0.75)training_data <- training(gap_split) testing_data <- testing(gap_split)nrow(training_data)
3003
nrow(testing_data)
1001
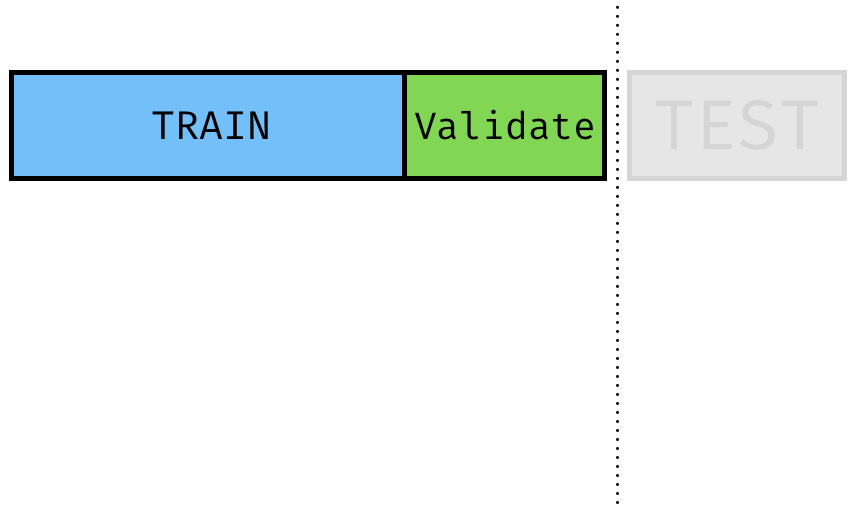
Train-Validate Split
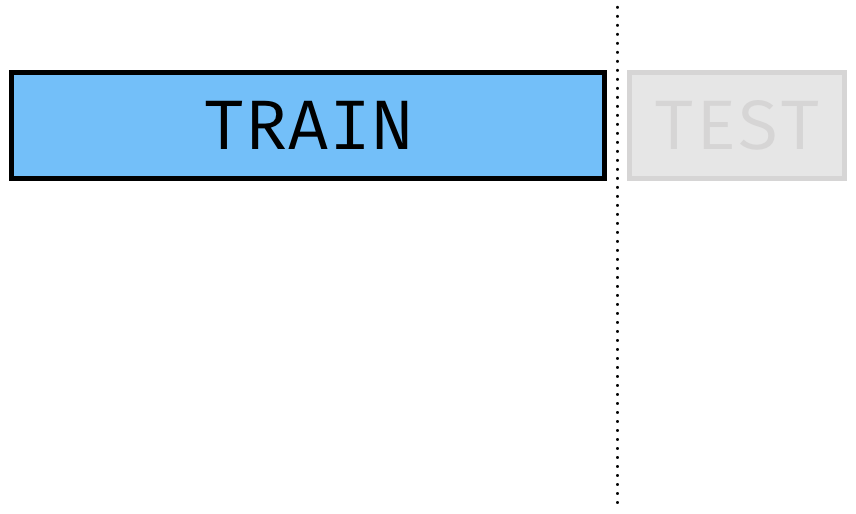
Train-Validate Split
Cross Validation
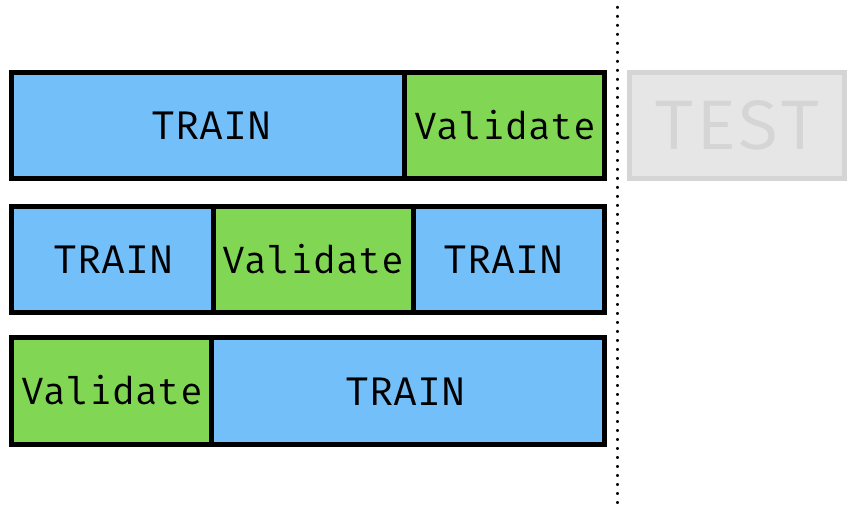
vfold_cv()
library(rsample) cv_split <- vfold_cv(training_data, v = 3)cv_split
# 3-fold cross-validation
# A tibble: 3 x 2
splits id
<list> <chr>
1 <S3: rsplit> Fold1
2 <S3: rsplit> Fold2
3 <S3: rsplit> Fold3
Mapping train & validate
cv_data <- cv_split %>%
mutate(train = map(splits, ~training(.x)),
validate = map(splits, ~testing(.x)))
Cross Validated Models
head(cv_data)
# A tibble: 3 x 4
splits id train validate
* <list> <chr> <list> <list>
1 <S3: rsplit> Fold1 <tibble [2,002 × 7]> <tibble [1,001 × 7]>
2 <S3: rsplit> Fold2 <tibble [2,002 × 7]> <tibble [1,001 × 7]>
3 <S3: rsplit> Fold3 <tibble [2,002 × 7]> <tibble [1,001 × 7]>
cv_models_lm <- cv_data %>%
mutate(model = map(train, ~lm(formula = life_expectancy~., data = .x)))
Let's practice!
Machine Learning in the Tidyverse

