Uniform Manifold Approximation and Projection (UMAP)
Dimensionality Reduction in R

Matt Pickard
Owner, Pickard Predictives, LLC
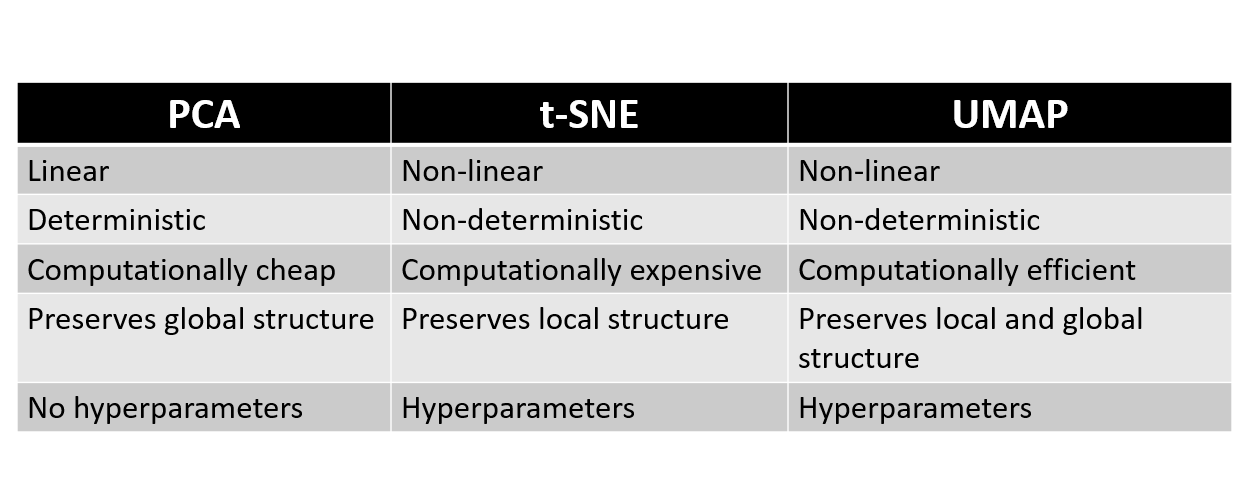
PCA, t-SNE, and UMAP
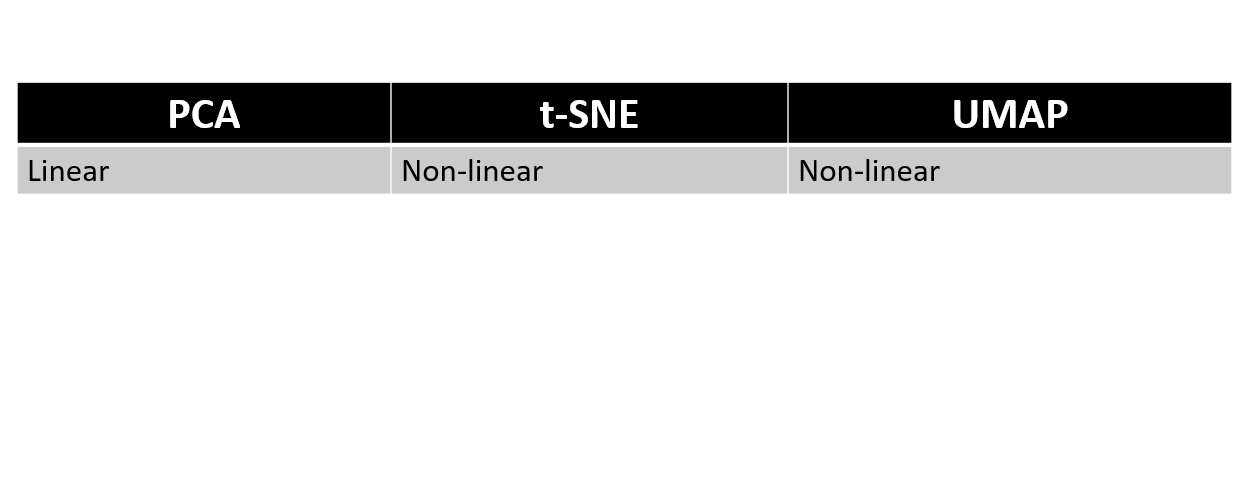
PCA, t-SNE, and UMAP
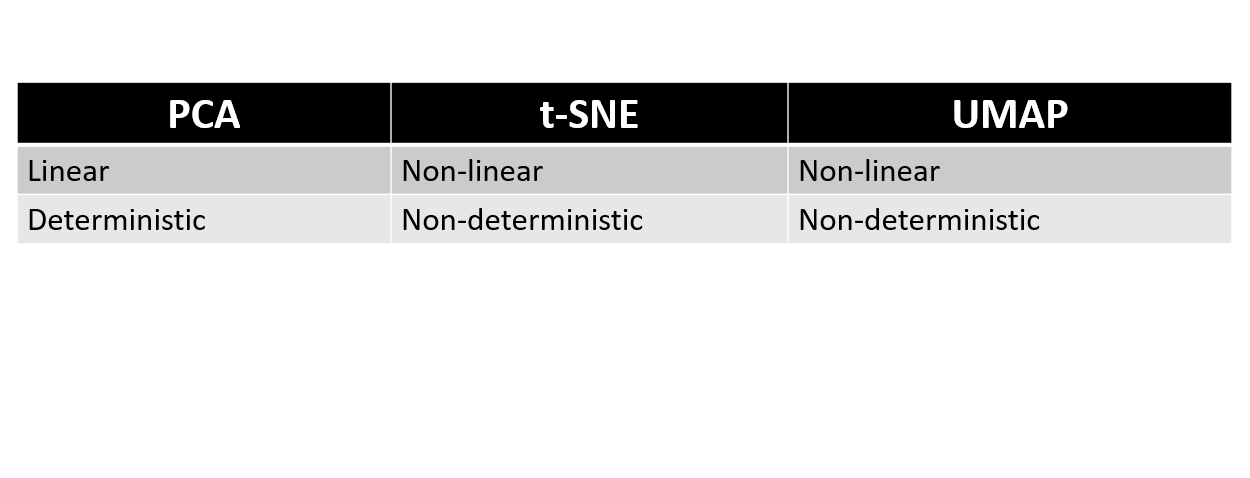
PCA, t-SNE, and UMAP
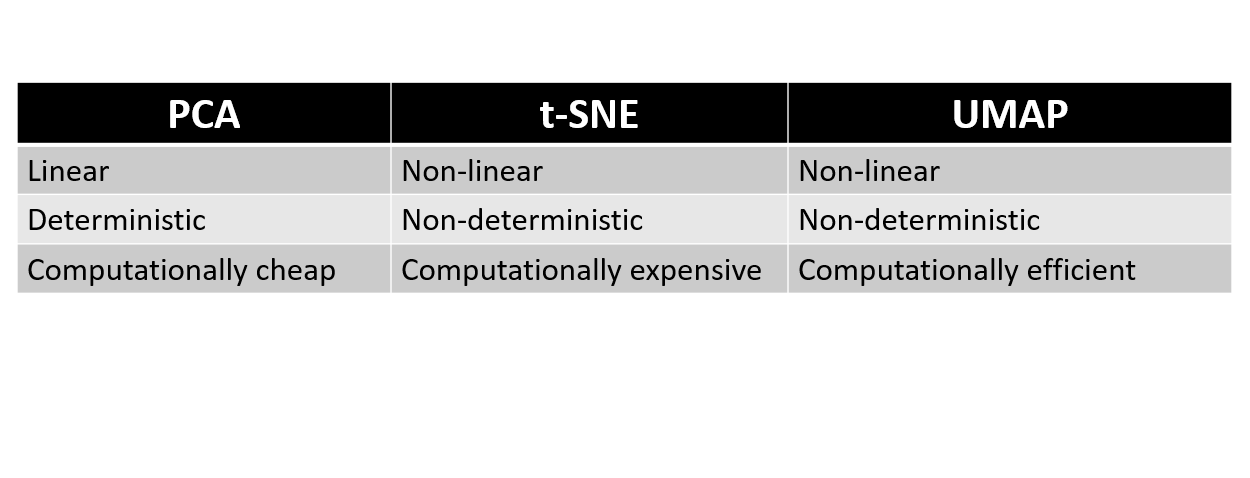
PCA, t-SNE, and UMAP
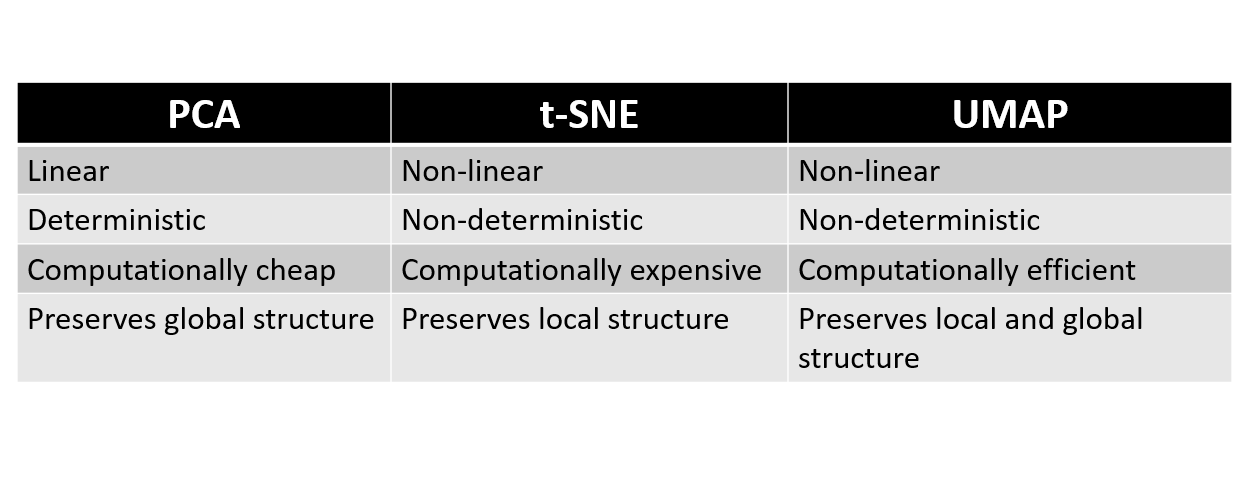
PCA, t-SNE, and UMAP
UMAP has similar hyperparameters that can be tuned.
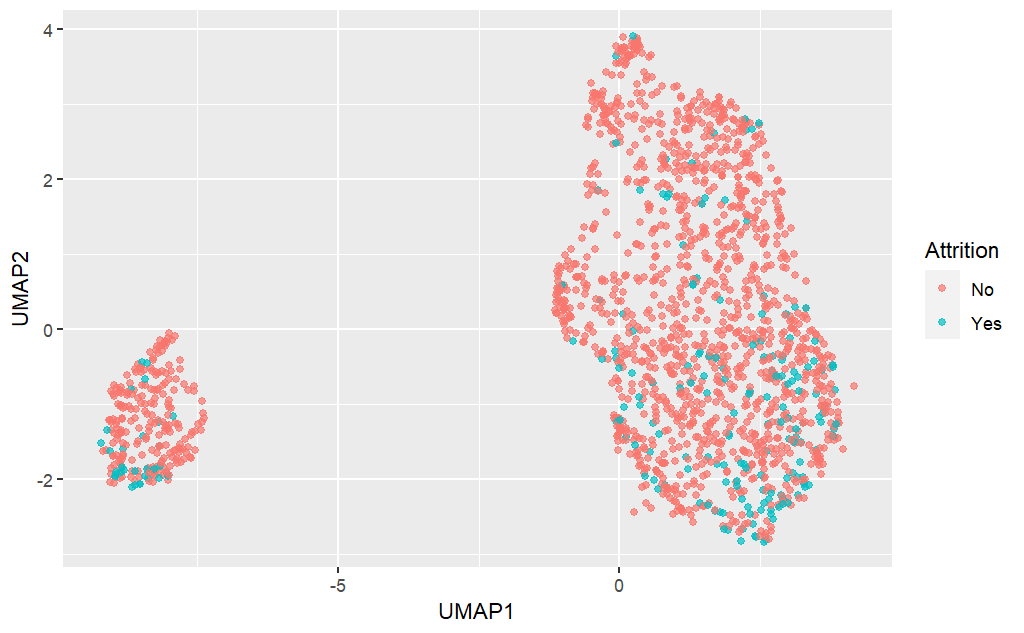
UMAP plot
library(embed)set.seed(1234) umap_df <- recipe(Attrition ~ ., data = attrition_df) %>% step_normalize(all_predictors()) %>% step_umap(all_predictors(), num_comp = 2) %>% prep() %>% juice()umap_df %>% ggplot(aes(x = UMAP1, y = UMAP2, color = Attrition)) + geom_point(alpha = 0.7)
UMAP: employee attrition
UMAP in tidymodels
Create recipe
umap_recipe <- recipe(Attrition ~ ., data = train) %>%
step_normalize(all_predictors()) %>%
step_umap(all_predictors(), num_comp = 4)
Create model spec
umap_lr_model <- linear_reg()
UMAP in tidymodels
Create workflow
umap_lr_workflow <- workflow() %>%
add_recipe(umap_recipe) %>%
add_model(umap_lr_model)
Fit the workflow
umap_lr_fit <- umap_lr_workflow %>%
fit(data = train)
UMAP in tidymodels
Evaluate the model
predict_umap_df <- test %>%
bind_cols(predict = predict(umap_lr_fit, test))
rmse(predict_umap_df, Attrition, .pred_class)
Let's practice!
Dimensionality Reduction in R

