Interpreting and visualising PCA models with factoextra
Dimensionality Reduction in R

Alexandros Tantos
Assistant Professor, Aristotle University of Thessaloniki
Plotting contributions of variables
fviz_pca_var(mtcars_pca,
col.var = "contrib",
gradient.cols = c("#bb2e00", "#002bbb"),
repel = TRUE)
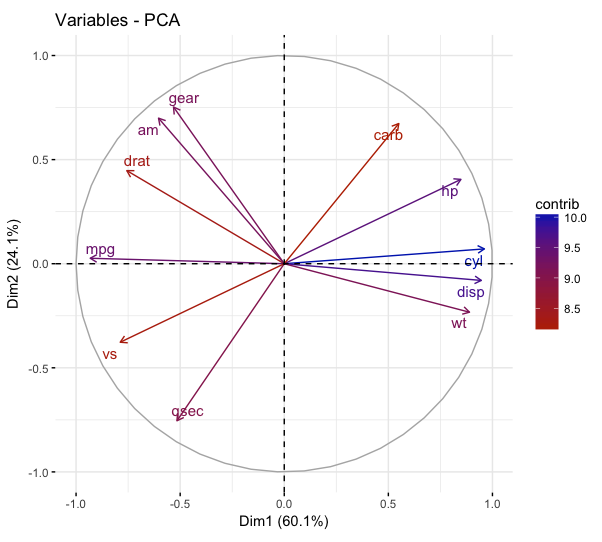
Plotting contributions of selected variables
fviz_pca_var(mtcars_pca,
select.var = list(contrib = 4),
repel = TRUE)
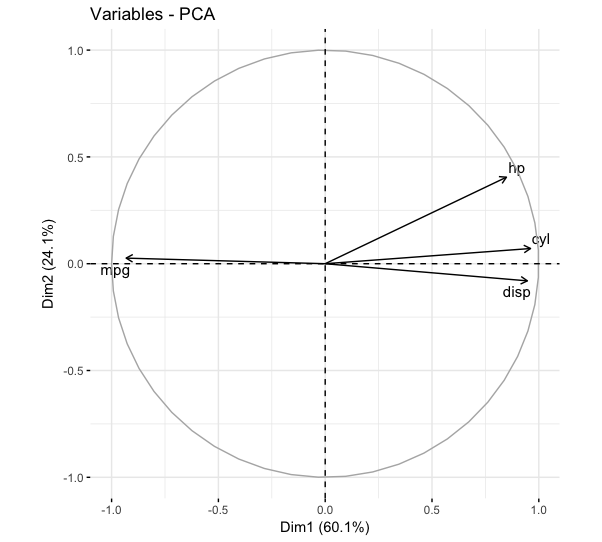
Barplotting the contributions of variables
fviz_contrib(mtcars_pca,
choice = "var",
axes = 1,
top = 5)
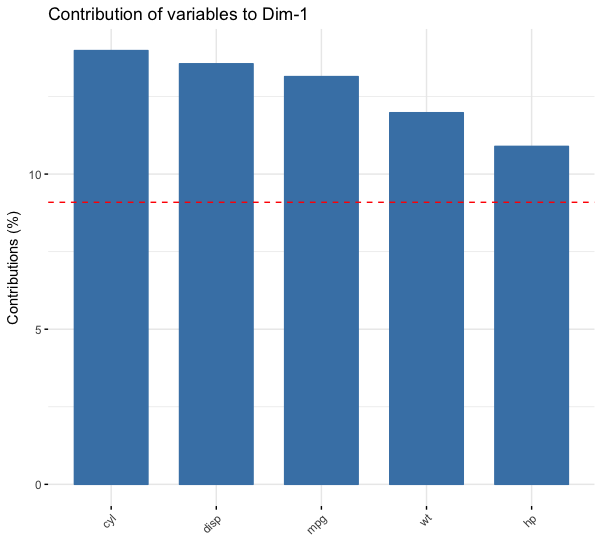
Plotting cos2 for individuals
fviz_pca_ind(mtcars_pca,
col.ind="cos2",
gradient.cols = c("#bb2e00", "#002bbb"),
repel = TRUE)
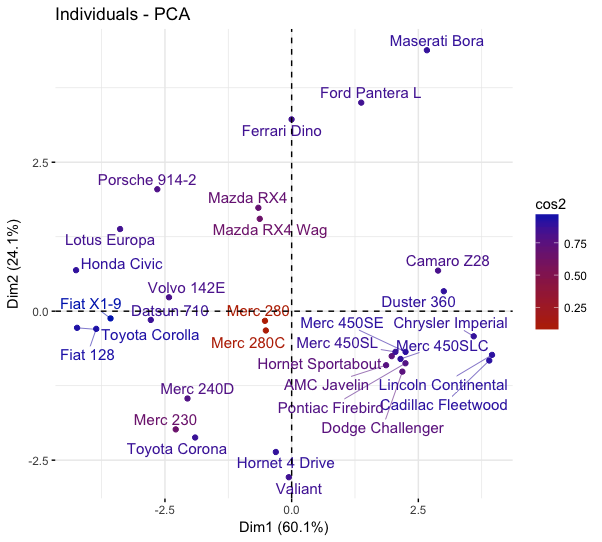
Plotting cos2 for selected individuals
fviz_pca_ind(mtcars_pca,
select.ind = list(cos2 = 0.8),
gradient.cols = c("#bb2e00", "#002bbb"),
repel = TRUE)
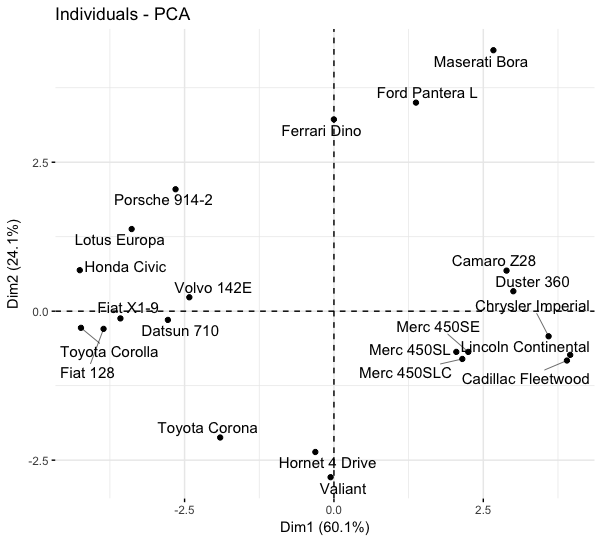
Barplotting cos2 for individuals
fviz_cos2(mtcars_pca,
choice = "ind",
axes = 1,
top = 10)
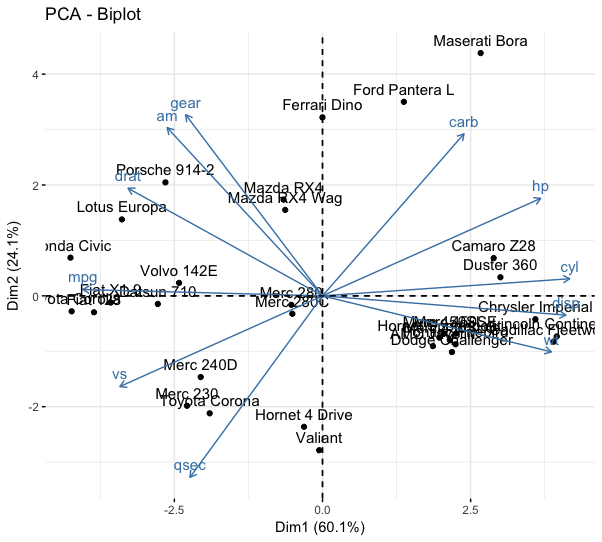
Biplots
fviz_pca_biplot(mtcars_pca)
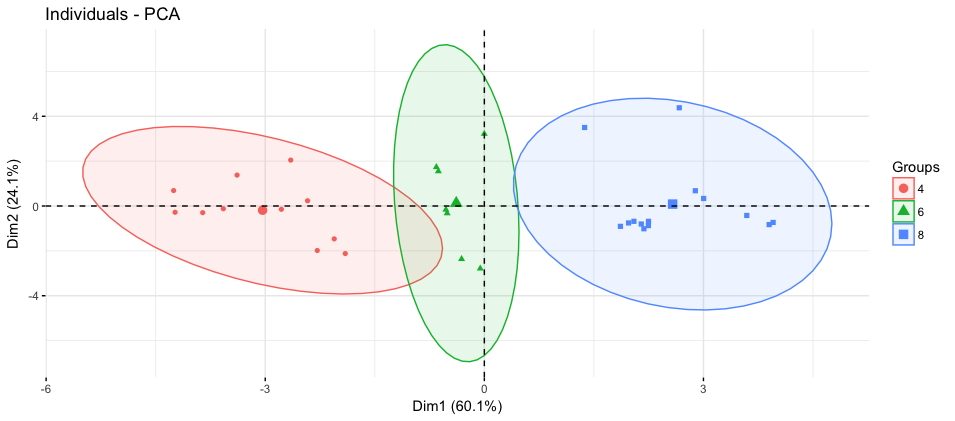
Adding ellipsoids
mtcars$cyl <- as.factor(mtcars$cyl)
fviz_pca_ind(mtcars_pca,
label="var",
habillage=mtcars$cyl,
addEllipses=TRUE)
Let's practice!
Dimensionality Reduction in R

